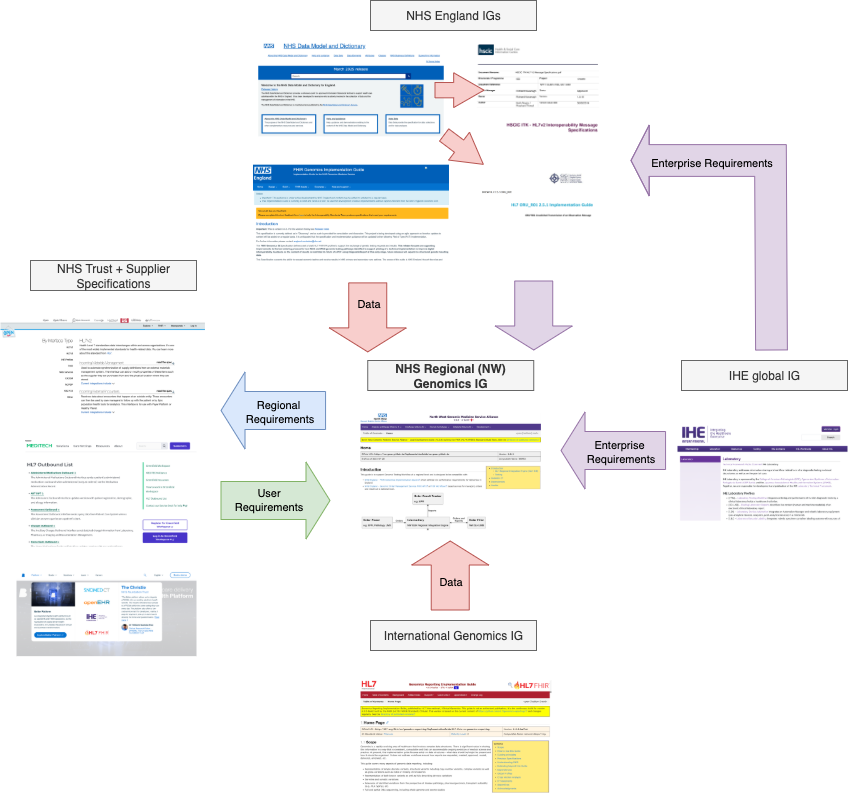
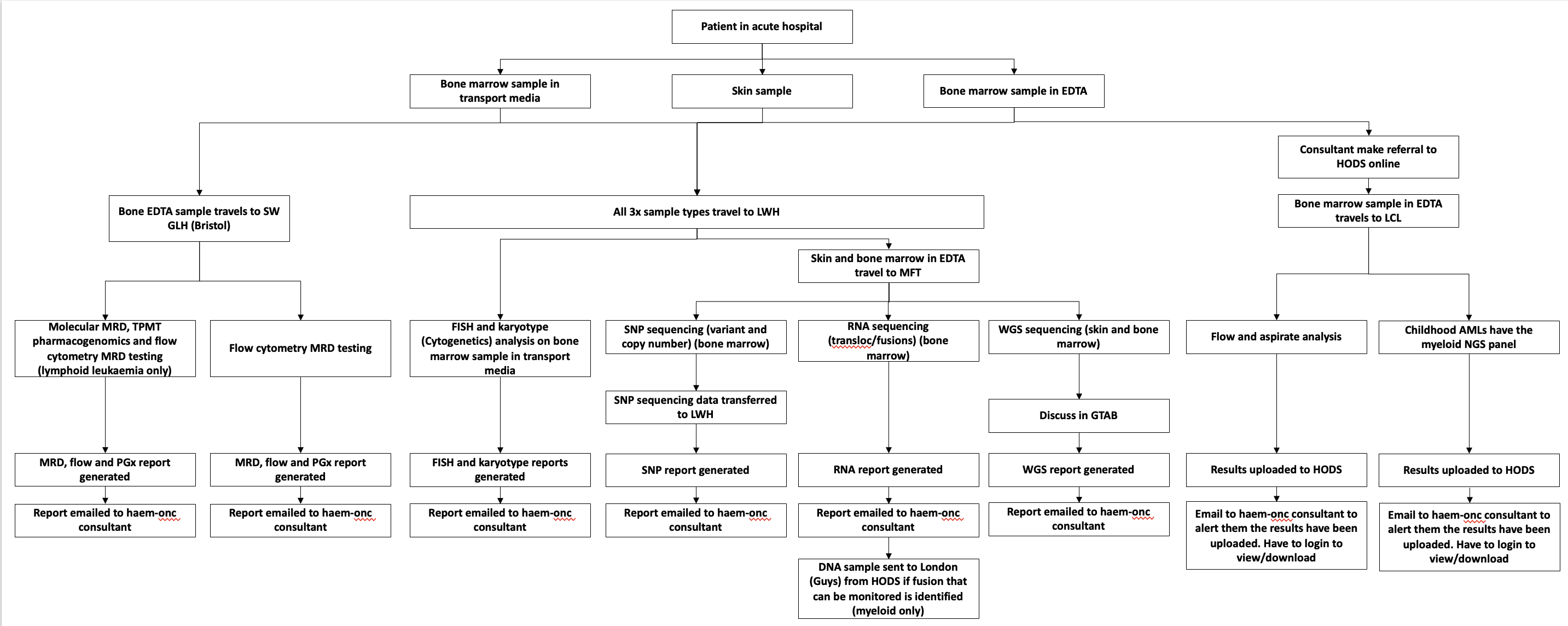
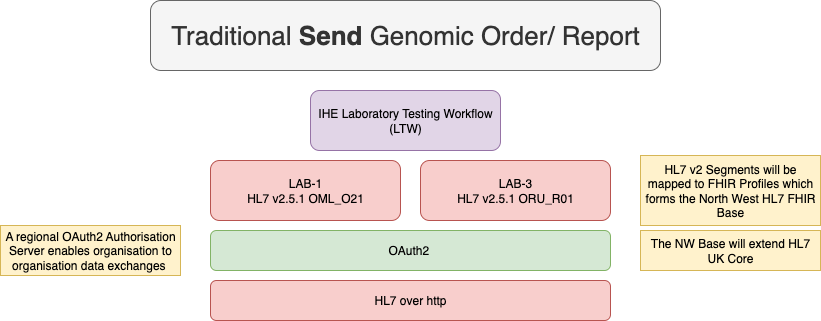
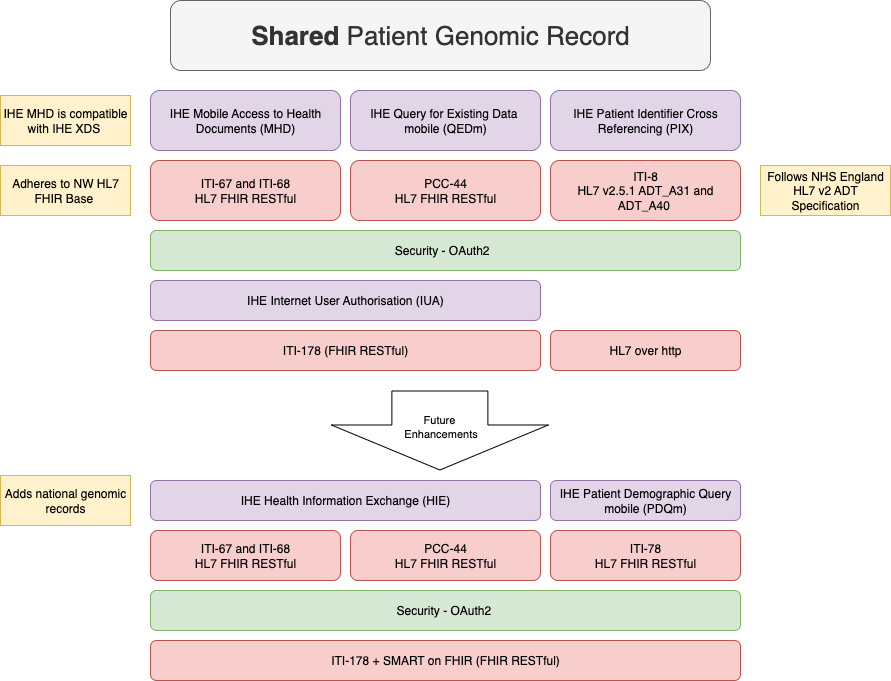
|
Type
|
Reference
|
Content
|
|
web
|
en.wikipedia.org
|
HL7 v3 Clinical Document Architecture
|
|
web
|
specifications.openehr.org
|
openEHR
|
|
web
|
profiles.ihe.net
|
Recommend IHE-IUA
|
|
web
|
browser.ihtsdotools.org
|
Mapping from (not specified) to All Codes SCT ValueSet
|
|
web
|
browser.ihtsdotools.org
|
Mapping from All Codes SCT ValueSet
to All Codes SCT ValueSet
|
|
web
|
www.nwgenomics.nhs.uk
|
contact
: https://www.nwgenomics.nhs.uk/contact-us
|
|
web
|
hl7.eu
|
Regional Hl7 v2 MDM_T02
The format of the report is PDF, in future for England/EU document sharing the format may change to HL7 EU Laboratory Report
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Event
Message
|
|
web
|
drive.google.com
|
NHS England HL7 v2 ADT Message Specification
|
|
web
|
profiles.ihe.net
|
Related to IHE Patient Encounter Management [ITI-31]
|
|
web
|
drive.google.com
|
NHS England HL7 v2 ADT Message Specification
|
|
web
|
hl7-definition.caristix.com
|
Create Patient HL7 v2.4 ADT_28
|
|
web
|
hl7-definition.caristix.com
|
Update Patient HL7 v2.4 ADT_31
|
|
web
|
hl7-definition.caristix.com
|
Update (merge) Patient HL7 v2.4 ADT_40
|
|
web
|
profiles.ihe.net
|
Related to IHE Patient Identifier Cross-referencing for mobile (PIXm)
|
|
web
|
snomed.info
|
77386006
|
|
web
|
snomed.info
|
255407002
|
|
web
|
snomed.info
|
721981007
|
|
web
|
snomed.info
|
721963009
|
|
web
|
snomed.info
|
734163000
|
|
web
|
snomed.info
|
371525003
|
|
web
|
snomed.info
|
422735006
|
|
web
|
snomed.info
|
371537001
|
|
web
|
snomed.info
|
419891008
|
|
web
|
snomed.info
|
4241000179101
|
|
web
|
snomed.info
|
721965002
|
|
web
|
snomed.info
|
163221000000102
|
|
web
|
snomed.info
|
927061000000101
|
|
web
|
snomed.info
|
149671000000105
|
|
web
|
snomed.info
|
820161000000108
|
|
web
|
snomed.info
|
820191000000102
|
|
web
|
snomed.info
|
824231000000100
|
|
web
|
snomed.info
|
820211000000103
|
|
web
|
snomed.info
|
823611000000107
|
|
web
|
snomed.info
|
820441000000103
|
|
web
|
snomed.info
|
820221000000109
|
|
web
|
snomed.info
|
829201000000105
|
|
web
|
snomed.info
|
820451000000100
|
|
web
|
snomed.info
|
820461000000102
|
|
web
|
snomed.info
|
820471000000109
|
|
web
|
snomed.info
|
823631000000104
|
|
web
|
snomed.info
|
820481000000106
|
|
web
|
snomed.info
|
820491000000108
|
|
web
|
snomed.info
|
823641000000108
|
|
web
|
snomed.info
|
24681000000104
|
|
web
|
snomed.info
|
824321000000109
|
|
web
|
snomed.info
|
823651000000106
|
|
web
|
snomed.info
|
823681000000100
|
|
web
|
snomed.info
|
823691000000103
|
|
web
|
snomed.info
|
823701000000103
|
|
web
|
snomed.info
|
824331000000106
|
|
web
|
snomed.info
|
824341000000102
|
|
web
|
snomed.info
|
25611000000107
|
|
web
|
snomed.info
|
823721000000107
|
|
web
|
snomed.info
|
823761000000104
|
|
web
|
snomed.info
|
909921000000109
|
|
web
|
snomed.info
|
823781000000108
|
|
web
|
snomed.info
|
1325651000000108
|
|
web
|
snomed.info
|
820241000000102
|
|
web
|
snomed.info
|
962381000000101
|
|
web
|
snomed.info
|
1219151000000104
|
|
web
|
snomed.info
|
25831000000103
|
|
web
|
snomed.info
|
416779005
|
|
web
|
snomed.info
|
820501000000102
|
|
web
|
snomed.info
|
820511000000100
|
|
web
|
snomed.info
|
820251000000104
|
|
web
|
snomed.info
|
823831000000103
|
|
web
|
snomed.info
|
823661000000109
|
|
web
|
snomed.info
|
823841000000107
|
|
web
|
snomed.info
|
725869001
|
|
web
|
snomed.info
|
1129271000000109
|
|
web
|
snomed.info
|
1363671000000109
|
|
web
|
snomed.info
|
1198111000000109
|
|
web
|
snomed.info
|
1076911000000107
|
|
web
|
snomed.info
|
4321000179101
|
|
web
|
snomed.info
|
1054291000000102
|
|
web
|
snomed.info
|
1054281000000104
|
|
web
|
snomed.info
|
4341000179107
|
|
web
|
snomed.info
|
1054181000000105
|
|
web
|
snomed.info
|
909871000000100
|
|
web
|
snomed.info
|
1054161000000101
|
|
web
|
snomed.info
|
824781000000106
|
|
web
|
snomed.info
|
824791000000108
|
|
web
|
snomed.info
|
820271000000108
|
|
web
|
snomed.info
|
1099461000000101
|
|
web
|
snomed.info
|
1325821000000102
|
|
web
|
snomed.info
|
185361000000102
|
|
web
|
snomed.info
|
826501000000100
|
|
web
|
snomed.info
|
824801000000107
|
|
web
|
snomed.info
|
24761000000103
|
|
web
|
snomed.info
|
823871000000101
|
|
web
|
snomed.info
|
229054004
|
|
web
|
snomed.info
|
824831000000101
|
|
web
|
snomed.info
|
307930005
|
|
web
|
snomed.info
|
826511000000103
|
|
web
|
snomed.info
|
820291000000107
|
|
web
|
snomed.info
|
826621000000105
|
|
web
|
snomed.info
|
826631000000107
|
|
web
|
snomed.info
|
826521000000109
|
|
web
|
snomed.info
|
308575004
|
|
web
|
snomed.info
|
310854009
|
|
web
|
snomed.info
|
308619006
|
|
web
|
snomed.info
|
270372007
|
|
web
|
snomed.info
|
270370004
|
|
web
|
snomed.info
|
307881004
|
|
web
|
snomed.info
|
270358003
|
|
web
|
snomed.info
|
308621001
|
|
web
|
snomed.info
|
820301000000106
|
|
web
|
snomed.info
|
823951000000100
|
|
web
|
snomed.info
|
826541000000102
|
|
web
|
snomed.info
|
826651000000100
|
|
web
|
snomed.info
|
408403008
|
|
web
|
snomed.info
|
823881000000104
|
|
web
|
snomed.info
|
827701000000106
|
|
web
|
snomed.info
|
772790007
|
|
web
|
snomed.info
|
822761000000108
|
|
web
|
snomed.info
|
1054091000000108
|
|
web
|
snomed.info
|
1054071000000109
|
|
web
|
snomed.info
|
827711000000108
|
|
web
|
snomed.info
|
822771000000101
|
|
web
|
snomed.info
|
822781000000104
|
|
web
|
snomed.info
|
822801000000103
|
|
web
|
snomed.info
|
822791000000102
|
|
web
|
snomed.info
|
827721000000102
|
|
web
|
snomed.info
|
822811000000101
|
|
web
|
snomed.info
|
824861000000106
|
|
web
|
snomed.info
|
371527006
|
|
web
|
snomed.info
|
822821000000107
|
|
web
|
snomed.info
|
822831000000109
|
|
web
|
snomed.info
|
823891000000102
|
|
web
|
snomed.info
|
962401000000101
|
|
web
|
snomed.info
|
4271000179106
|
|
web
|
snomed.info
|
983701000000101
|
|
web
|
snomed.info
|
725870000
|
|
web
|
snomed.info
|
725956001
|
|
web
|
snomed.info
|
770573007
|
|
web
|
snomed.info
|
770574001
|
|
web
|
snomed.info
|
770572002
|
|
web
|
snomed.info
|
770575000
|
|
web
|
snomed.info
|
782660000
|
|
web
|
snomed.info
|
782659005
|
|
web
|
snomed.info
|
1129261000000102
|
|
web
|
snomed.info
|
1129251000000100
|
|
web
|
snomed.info
|
1129241000000103
|
|
web
|
snomed.info
|
1129231000000107
|
|
web
|
snomed.info
|
4201000179104
|
|
web
|
snomed.info
|
1219161000000101
|
|
web
|
snomed.info
|
149691000000109
|
|
web
|
snomed.info
|
229059009
|
|
web
|
snomed.info
|
823901000000101
|
|
web
|
snomed.info
|
823931000000107
|
|
web
|
snomed.info
|
823941000000103
|
|
web
|
snomed.info
|
1186936003
|
|
web
|
snomed.info
|
119373006
|
|
web
|
snomed.info
|
309201001
|
|
web
|
snomed.info
|
258580003
|
|
web
|
snomed.info
|
122552005
|
|
web
|
snomed.info
|
122556008
|
|
web
|
snomed.info
|
737357006
|
|
web
|
snomed.info
|
122555007
|
|
web
|
snomed.info
|
440500007
|
|
web
|
snomed.info
|
119359002
|
|
web
|
snomed.info
|
733104004
|
|
web
|
snomed.info
|
258450006
|
|
web
|
snomed.info
|
258565009
|
|
web
|
snomed.info
|
702451000
|
|
web
|
snomed.info
|
309147000
|
|
web
|
snomed.info
|
258566005
|
|
web
|
snomed.info
|
441652008
|
|
web
|
snomed.info
|
3040001000004100
|
|
web
|
snomed.info
|
1003517007
|
|
web
|
snomed.info
|
122571007
|
|
web
|
snomed.info
|
418564007
|
|
web
|
snomed.info
|
119342007
|
|
web
|
snomed.info
|
441479001
|
|
web
|
snomed.info
|
16214131000119100
|
|
web
|
snomed.info
|
122575003
|
|
web
|
snomed.info
|
441673008
|
|
web
|
www.datadictionary.nhs.uk
|
General Medical Council Consultant Code. For more details see: NHS Data Model Dictionary
|
|
web
|
www.datadictionary.nhs.uk
|
General Practitioner (GMP) number. For more details see: NHS Data Model Dictionary
|
|
web
|
www.datadictionary.nhs.uk
|
NHS Number allocated to the patient in England and Wales. For more details see: NHS Data Model Dictionary
|
|
web
|
www.nw-gmsa.nhs.uk
|

|
|
web
|
www.nwgenomics.nhs.uk
|
IG © 2024+ NHS North West Genomics
. Package fhir.nwgenomics.nhs.uk#0.1.0 based on FHIR 4.0.1
. Generated 2026-02-04
Links: Table of Contents |
QA Report
| New Issue | Issues
Version History |
 |
Propose a change |
Propose a change
|
|
web
|
github.com
|
Links: Table of Contents
|
QA Report
| New Issue
| Issues
Version History
|
 |
Propose a change
|
Propose a change
|
|
web
|
github.com
|
Links: Table of Contents
|
QA Report
| New Issue
| Issues
Version History
|
 |
Propose a change
|
Propose a change
|
|
web
|
profiles.ihe.net
|
Links: Table of Contents
|
QA Report
| New Issue
| Issues
Version History
|
 |
Propose a change
|
Propose a change
|
|
web
|
www.ihe.net
|
Links: Table of Contents
|
QA Report
| New Issue
| Issues
Version History
|
 |
Propose a change
|
Propose a change
|
|
web
|
digital.nhs.uk
|
Manages the Order. This is the function of Genomic Order Management Service
and/or GLH LIMS
|
|
web
|
www.en13606.org
|
From ISO-13606
The set of information committed to one EHR as a result of a clinical encounter or a record documentation session. Examples of COMPOSITION are Progress note, Laboratory test result form, Radiology report, Referral letter, Clinic visit, Clinic letter, Discharge summary, Functional health assessment, Diabetes review.
|
|
web
|
profiles.ihe.net
|
IHE Basic Patient Privacy Consents (BPPC)
|
|
web
|
profiles.ihe.net
|
IHE Privacy Consent on FHIR (PCF)
|
|
web
|
bmjopenquality.bmj.com
|
Implementation of Digital Consent at Sandwell and West Birmingham NHS Trust: A Quality Improvement Project
|
|
web
|
mft.nhs.uk
|
North West Genomics - Consent for a Genomic Test
|
|
web
|
editor.swagger.io
|
The OpenAPISwagger Definition file
below, can be viewed using Swagger Editor
|
|
web
|
www.england.nhs.uk
|
Genomic Test Directory
|
|
web
|
www.england.nhs.uk
|
Test Outcome Code from NHS England Genomics Test Reporting Specification
|
|
web
|
confluence.ihtsdotools.org
|
For Implementation Guide see SNOMED CT Implementation Guide for the LOINC Ontology
|
|
web
|
en.wikipedia.org
|
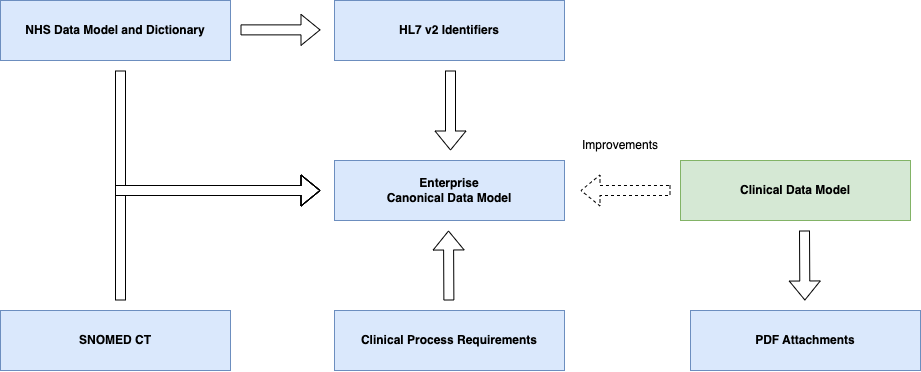
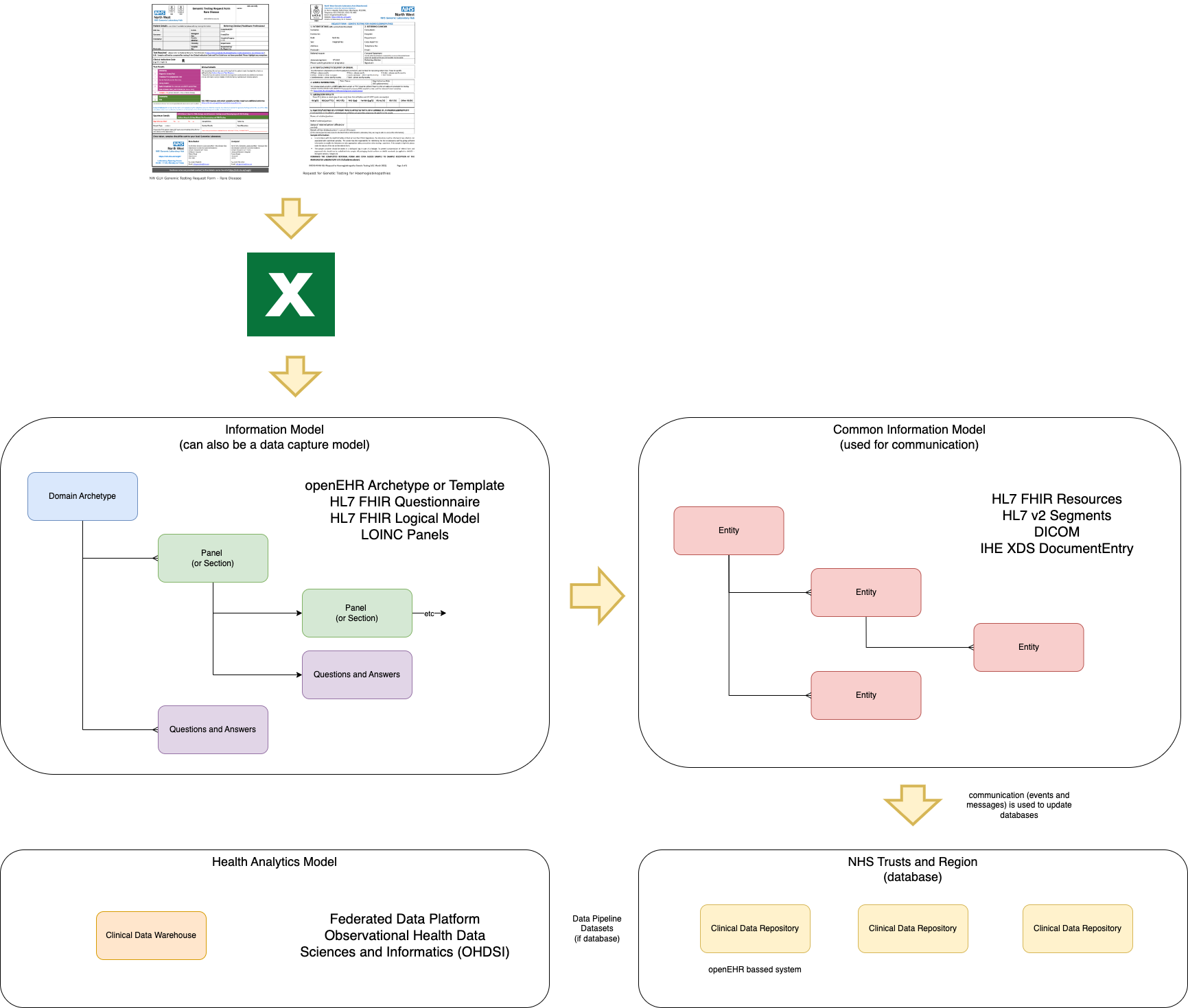
Domain Archetype
is the result of a collaboration process which aims to form a series of common archetypes within a domain (the domain in this case is Genomics).
The initial format for this archetype is often a spreadsheet that is expressed in an electronic format. The format we are using in this guide is HL7 FHIR Questionnaire
, other formats can be used such as openEHR Archetype (see Genomic Variant Result
)
|
|
web
|
ckm.openehr.org
|
Domain Archetype
is the result of a collaboration process which aims to form a series of common archetypes within a domain (the domain in this case is Genomics).
The initial format for this archetype is often a spreadsheet that is expressed in an electronic format. The format we are using in this guide is HL7 FHIR Questionnaire
, other formats can be used such as openEHR Archetype (see Genomic Variant Result
)
|
|
web
|
en.wikipedia.org
|
The domain archetypes are implemented via a Canonical Data Model
, which is common across all technical formats (i.e. HL7 v2 and HL7 FHIR) and is described using HL7 FHIR.
|
|
web
|
www.datadictionary.nhs.uk
|
Elements from NHS England FHIR Genomics Implementation Guide
have been incorporated into this guide, in particular the use of NHS Data Model and Dictionary Model
identifiers and other identifiers already present in HL7 v2 OML and ORU.
See Identities and Codes
for more details.
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Both domain archetypes have a very strong focus on the use of Correlation Identifier (Enterprise Integration Patterns)
or Rule 3: Reference Other Aggregates by Identity (Implementing Domain Driven Design)
, which is also consistent with IHE/HL7 concepts of Bounded Context (martinfowler.com)
|
|
web
|
www.archi-lab.io
|
Both domain archetypes have a very strong focus on the use of Correlation Identifier (Enterprise Integration Patterns)
or Rule 3: Reference Other Aggregates by Identity (Implementing Domain Driven Design)
, which is also consistent with IHE/HL7 concepts of Bounded Context (martinfowler.com)
|
|
web
|
martinfowler.com
|
Both domain archetypes have a very strong focus on the use of Correlation Identifier (Enterprise Integration Patterns)
or Rule 3: Reference Other Aggregates by Identity (Implementing Domain Driven Design)
, which is also consistent with IHE/HL7 concepts of Bounded Context (martinfowler.com)
|
|
web
|
www.datadictionary.nhs.uk
|
This includes making use of FHIR Identifier assigner.identifier.value
(HL7 v2 Assigning Facility
in a variety of ID types) to distinguish these identifiers between different organisations, the recommendation is to use ODS Code
, e.g.
|
|
web
|
digital.nhs.uk
|
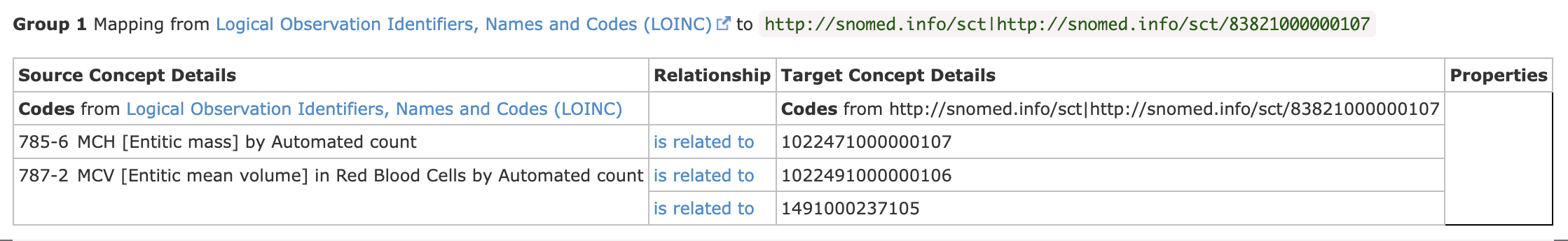
The coding (B0001, B0300, B0307, etc) is using local laboratory coding, ideally we want all organisations to communicate via standard coding and in the UK this preferred clinical coding is SNOMED CT UK Edition 83821000000107
and the preferred coding standard for units is UCUM
. To use local codes would mean 20+ organisation maintaining code mappings between all the different local codesystems, by using SNOMED (or LOINC) this means they only need to maintain mappings between local codes and SNOMED (or LOINC)
|
|
web
|
ucum.org
|
The coding (B0001, B0300, B0307, etc) is using local laboratory coding, ideally we want all organisations to communicate via standard coding and in the UK this preferred clinical coding is SNOMED CT UK Edition 83821000000107
and the preferred coding standard for units is UCUM
. To use local codes would mean 20+ organisation maintaining code mappings between all the different local codesystems, by using SNOMED (or LOINC) this means they only need to maintain mappings between local codes and SNOMED (or LOINC)
|
|
web
|
www.opencodelists.org
|
OpenSAFELY codelists
which contain many valuesets used in primary care.
|
|
web
|
pathlabs.rlbuht.nhs.uk
|
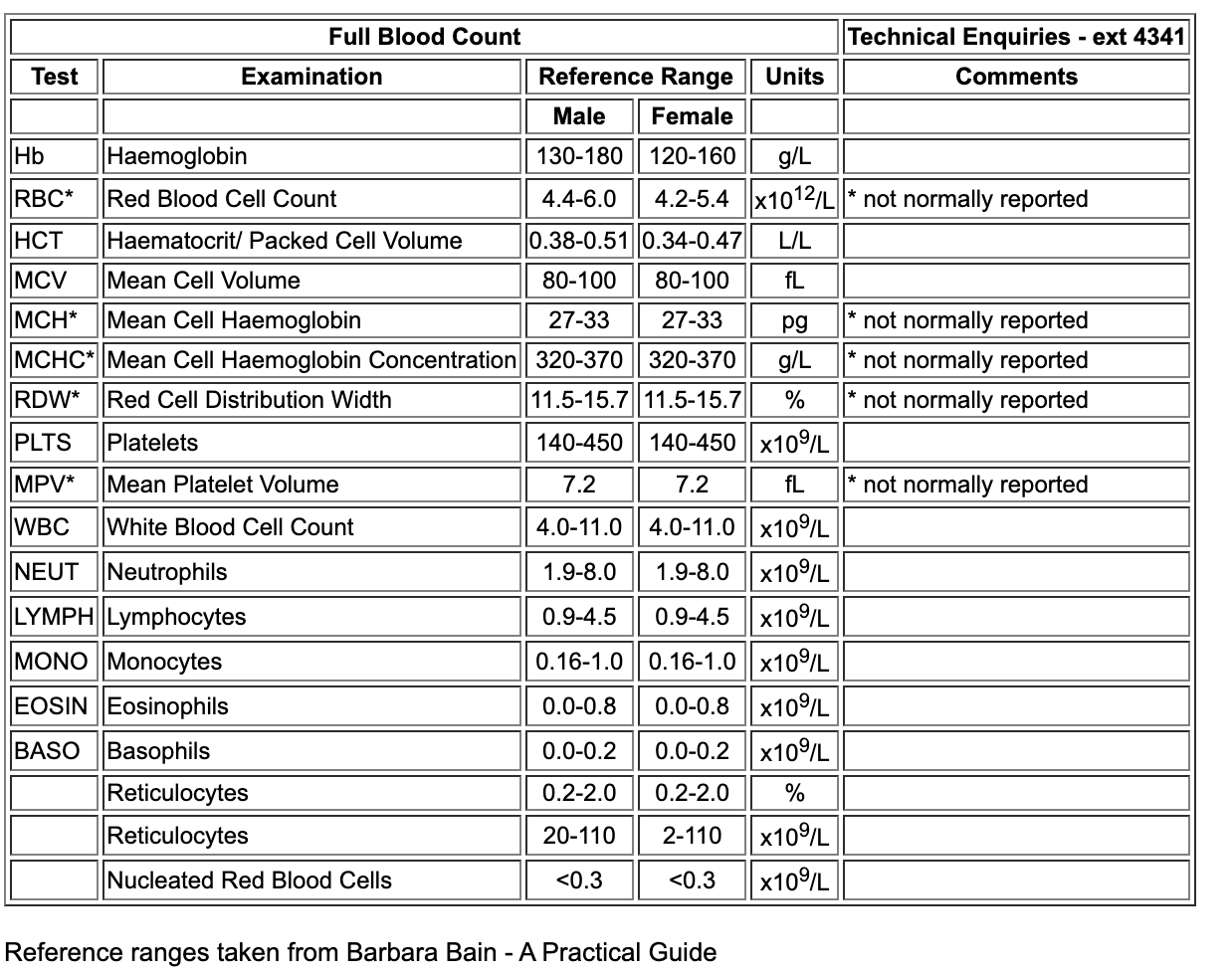
Internet searches also reveal several NHS Trusts providing documentation around Full Blood Count
, this often includes the local coding we saw with the HL7 v2 example. The example below is from University Hospitals of Liverpool Group - Full Blood Count
|
|
web
|
en.wikipedia.org
|
Code mappings should be created by a clinical coder
. In HL7 FHIR this code mapping is known as a ConceptMap
. NHS England has produced instructions for creating a ConceptMap
using the NHS England Ontology Server
|
|
web
|
digital.nhs.uk
|
Code mappings should be created by a clinical coder
. In HL7 FHIR this code mapping is known as a ConceptMap
. NHS England has produced instructions for creating a ConceptMap
using the NHS England Ontology Server
|
|
web
|
digital.nhs.uk
|
Code mappings should be created by a clinical coder
. In HL7 FHIR this code mapping is known as a ConceptMap
. NHS England has produced instructions for creating a ConceptMap
using the NHS England Ontology Server
|
|
web
|
snomed.info
|
147071010000102
|
|
web
|
snomed.info
|
249121010000107
|
|
web
|
snomed.info
|
168331010000106
|
|
web
|
snomed.info
|
558261010000109
|
|
web
|
snomed.info
|
613061010000108
|
|
web
|
snomed.info
|
612991010000106
|
|
web
|
snomed.info
|
613001010000107
|
|
web
|
snomed.info
|
613181010000104
|
|
web
|
snomed.info
|
218801010000106
|
|
web
|
snomed.info
|
528301010000103
|
|
web
|
digital.nhs.uk
|
express a preference towards SNOMED CT
|
|
web
|
digital.nhs.uk
|
The use of FHIR resources for data which is currently HL7 v2 CE Data Type may need to be questioned.
For example mapping of HL7 v2 ORC-16
is to ServiceRequest.requestCode
according to ServiceRquest - HL7 v2 Mapping
, this is a v2 CE to FHIR CodeableConcept conversion.
Curently this is stated as an implied ServiceRequest.requestReference
mapping (ISSUE link: NHS England Developer Community Genomics Order Management Service - FHIR ServiceRequest reason
)
|
|
web
|
developer.community.nhs.uk
|
The use of FHIR resources for data which is currently HL7 v2 CE Data Type may need to be questioned.
For example mapping of HL7 v2 ORC-16
is to ServiceRequest.requestCode
according to ServiceRquest - HL7 v2 Mapping
, this is a v2 CE to FHIR CodeableConcept conversion.
Curently this is stated as an implied ServiceRequest.requestReference
mapping (ISSUE link: NHS England Developer Community Genomics Order Management Service - FHIR ServiceRequest reason
)
|
|
web
|
profiles.ihe.net
|
IHE HIE White Paper
|
|
web
|
profiles.ihe.net
|
IHE Cross-Enterprise Document Sharing (XDS.b)
XML
|
|
web
|
profiles.ihe.net
|
IHE Mobile access to Health Documents (MHD)
HL7 FHIR
|
|
web
|
profiles.ihe.net
|
IHE Query for Existing Data Mobile (QEDm)
HL7 FHIR
|
|
web
|
fhir.epic.com
|
EPIC on FHIR
|
|
web
|
fhir.meditech.com
|
Meditech FHIR
|
|
web
|
docs.oracle.com
|
FHIR R4 APIs for Oracle Health Millennium Platform
|
|
web
|
profiles.ihe.net
|
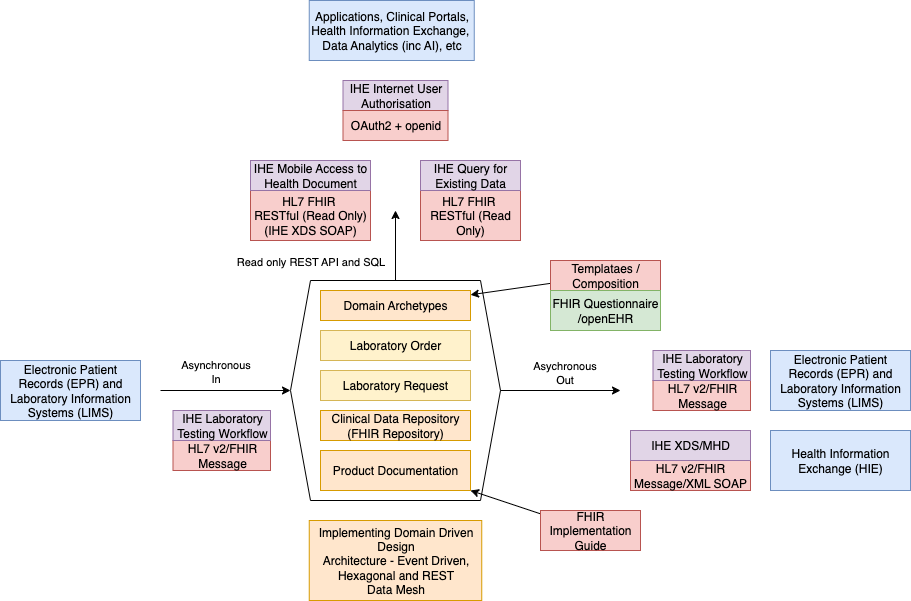
The Regional Clinical Data Repository (CDR) will adopt a similar FHIR RESTful approach to that used by Electronic Health Records (EHRs), and will also conform to IHE Query for Existing Data for Mobile (QEDm)
and IHE Mobile access to Health Documents (MHD)
|
|
web
|
digital.nhs.uk
|
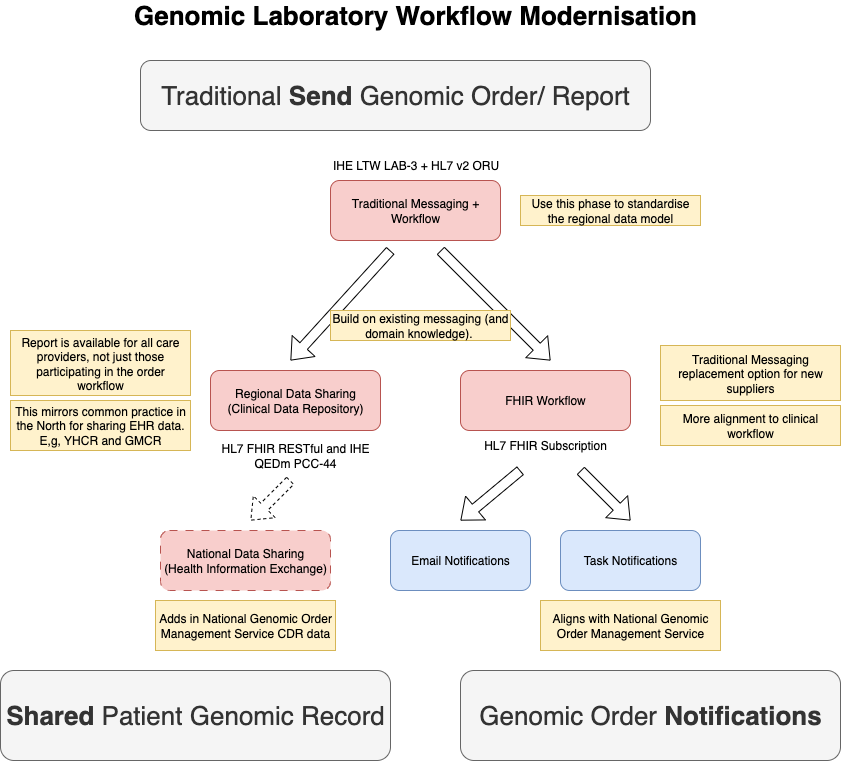
In the future, an alternative messaging approach using FHIR Subscription
and Event Notifications is expected to be supported.
The outline of this approach is shown below and is related to a similar approach used by NHS England Pathology FHIR specification
|
|
web
|
profiles.ihe.net
|
Provide and Register Document Set-b (ITI-41)
Provide Document Bundle (ITI-65)
Simplified Publish (ITI-105)
Generate Metadata (ITI-106)
|
|
web
|
profiles.ihe.net
|
Provide and Register Document Set-b (ITI-41)
Provide Document Bundle (ITI-65)
Simplified Publish (ITI-105)
Generate Metadata (ITI-106)
|
|
web
|
profiles.ihe.net
|
Provide and Register Document Set-b (ITI-41)
Provide Document Bundle (ITI-65)
Simplified Publish (ITI-105)
Generate Metadata (ITI-106)
|
|
web
|
profiles.ihe.net
|
Provide and Register Document Set-b (ITI-41)
Provide Document Bundle (ITI-65)
Simplified Publish (ITI-105)
Generate Metadata (ITI-106)
|
|
web
|
profiles.ihe.net
|
Registry Stored Query (ITI-18)
Find Document References (ITI-67)
|
|
web
|
profiles.ihe.net
|
Registry Stored Query (ITI-18)
Find Document References (ITI-67)
|
|
web
|
profiles.ihe.net
|
Retrieve Document Set (ITI-43)
Retrieve Document (ITI-68)
|
|
web
|
profiles.ihe.net
|
Retrieve Document Set (ITI-43)
Retrieve Document (ITI-68)
|
|
web
|
profiles.ihe.net
|
Mobile Query Existing Data (PCC-44)
|
|
web
|
hl7-definition.caristix.com
|
A. HL7 v2.5.1 MFN_M02 - Master files notification - Staff/practitioner master file
|
|
web
|
profiles.ihe.net
|
B. IHE Mobile Care Services Discovery (mCSD)
|
|
web
|
profiles.ihe.net
|
HL7 v2 MFN_M02 (or
IHE ITI-59
)
|
|
web
|
hl7-definition.caristix.com
|
The Care Service Update Source
sends a HL7 v2.5.1 MFN_M02 - Master files notification - Staff/practitioner master file
event to the RIE
|
|
web
|
digital.nhs.uk
|
Lookup up existing PractitionerRole via supplied identifiers (MRN or NHS Number) via a FHIR RESTful Search
.
|
|
web
|
digital.nhs.uk
|
If a PractitionerRole record is found, then update the Patient record via a FHIR RESTful Update
|
|
web
|
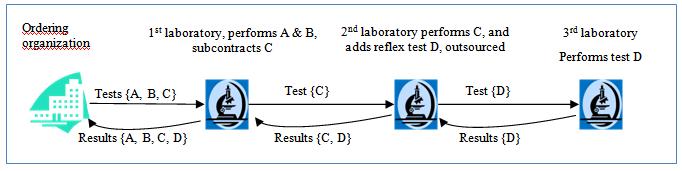
wiki.ihe.net
|
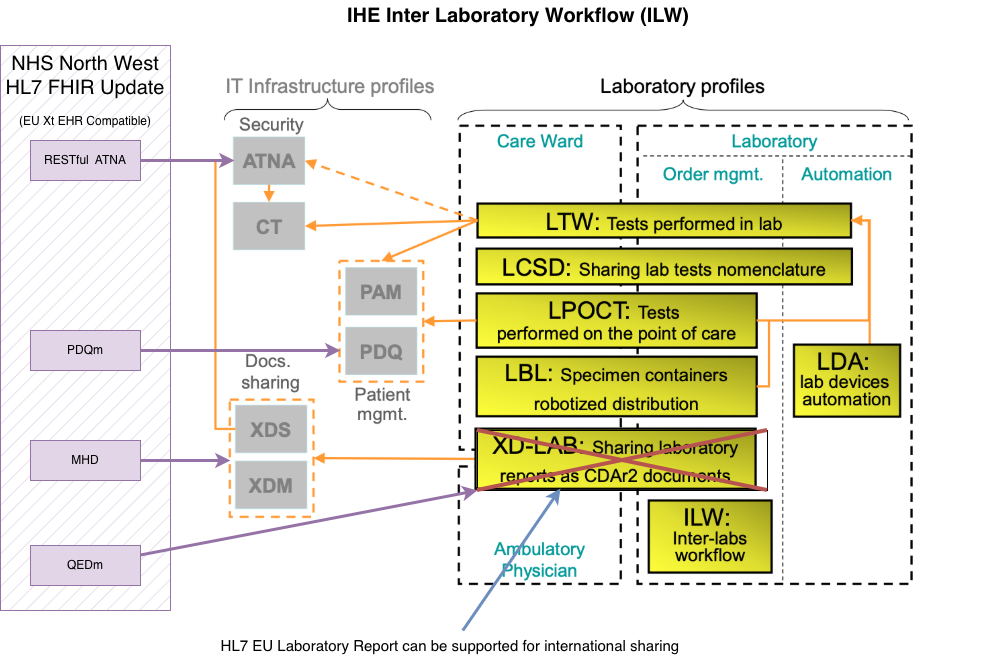
IHE Inter Laboratory Workflow
|
|
web
|
www.ihe.net
|
IHE Laboratory Technical Framework Supplement Inter-Laboratory Workflow (ILW)
|
|
web
|
digital.nhs.uk
|
NHS England - Genomic Order Management Service FHIR API
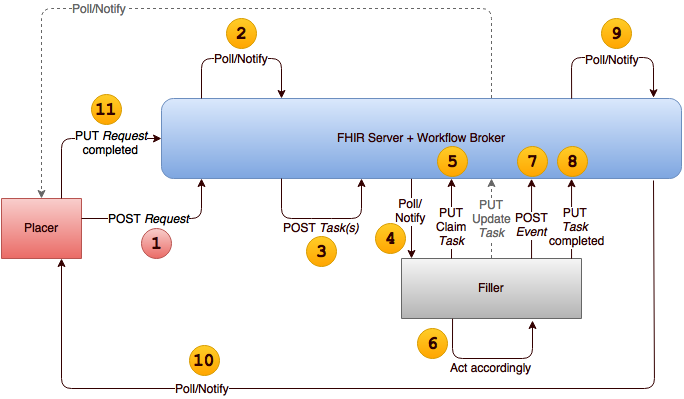
a FHIR Workflow
based service for managing orders and results at a national level.
|
|
web
|
profiles.ihe.net
|
IHE Internet User Authorization (IUA)
|
|
web
|
www.ihe.net
|
A. IHE Pathology and Laboratory Medicine (PaLM) Technical Framework - Volume 2a (PaLM TF-2a) Transactions
|
|
web
|
www.enterpriseintegrationpatterns.com
|
EIP - Command Message
|
|
web
|
martinfowler.com
|
The following messages are used to support creation and updating of the Genomics Test Order
aggregation
/ archetype
|
|
web
|
en.wikipedia.org
|
The following messages are used to support creation and updating of the Genomics Test Order
aggregation
/ archetype
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Command Message
|
|
web
|
www.enterpriseintegrationpatterns.com
|
EIP - Event Message
|
|
web
|
www.enterpriseintegrationpatterns.com
|
EIP - Document Message
|
|
web
|
www.rcr.ac.uk
|
The Royal College of Radiologists – Reporting networks - understanding the technical options
for ORC+OBR/FHIR DiagnosticReport
|
|
web
|
www.ihe-europe.net
|
IHE Europe Document Metadata
for OBX (value type = ED)/FHIR DocumentReference metadata
|
|
web
|
www.digihealthcare.scot
|
Digital Health and Care Scotland - (EH4001) CLINICAL DOCUMENT INDEXING STANDARDS
for OBX (value type = ED)/FHIR DocumentReference metadata
|
|
web
|
martinfowler.com
|
The following messages are used to support creation and updating of the Genomics Test Report
composition
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Document Message
|
|
web
|
www.enterpriseintegrationpatterns.com
|
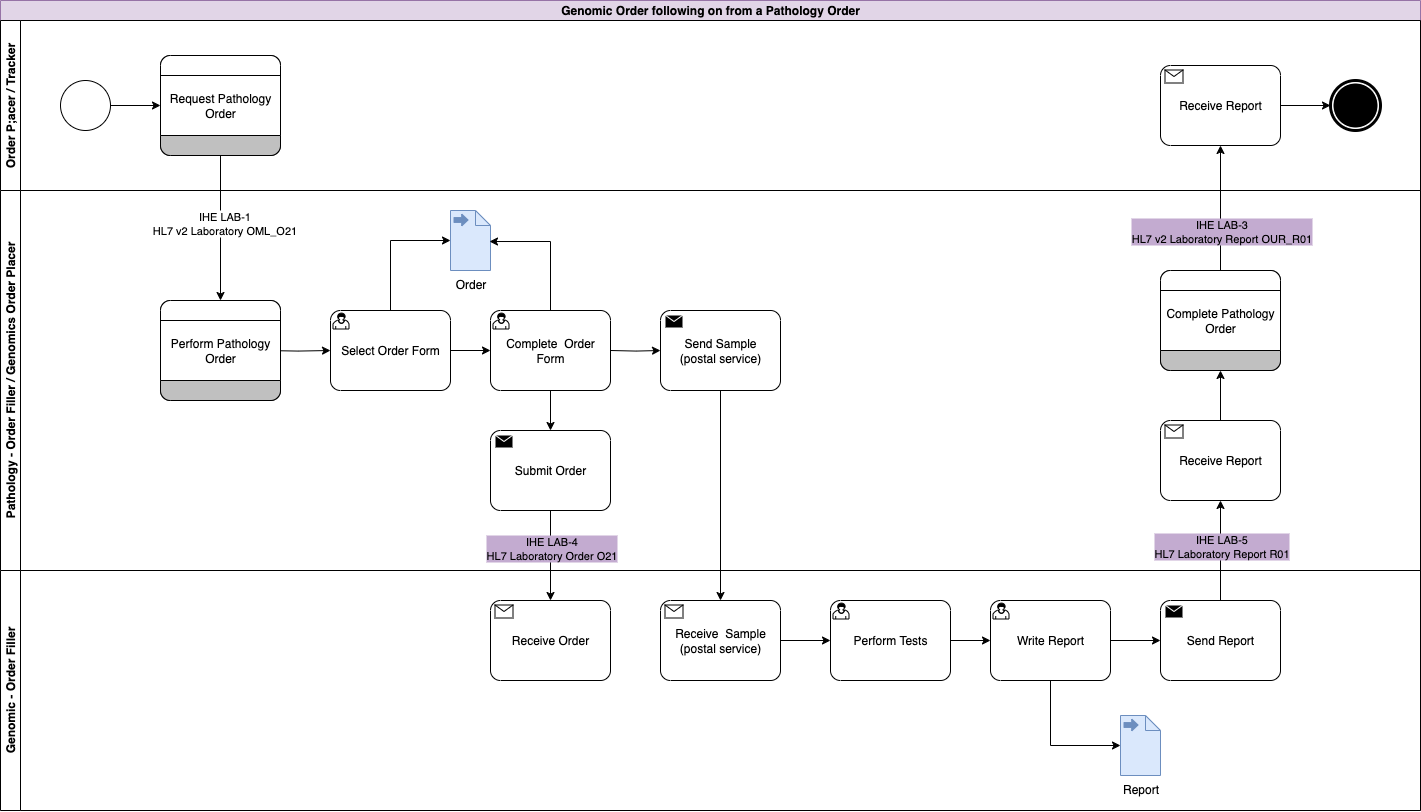
In this workflow, both the order and report are shared instead of being sent. The communication between the Order Placer and Filler changes to a conversation
rather than messaging pattern around the Fulfillment Task.
|
|
web
|
digital.nhs.uk
|
This modernisation is central to the NHS England Genomic Order Management Service FHIR API
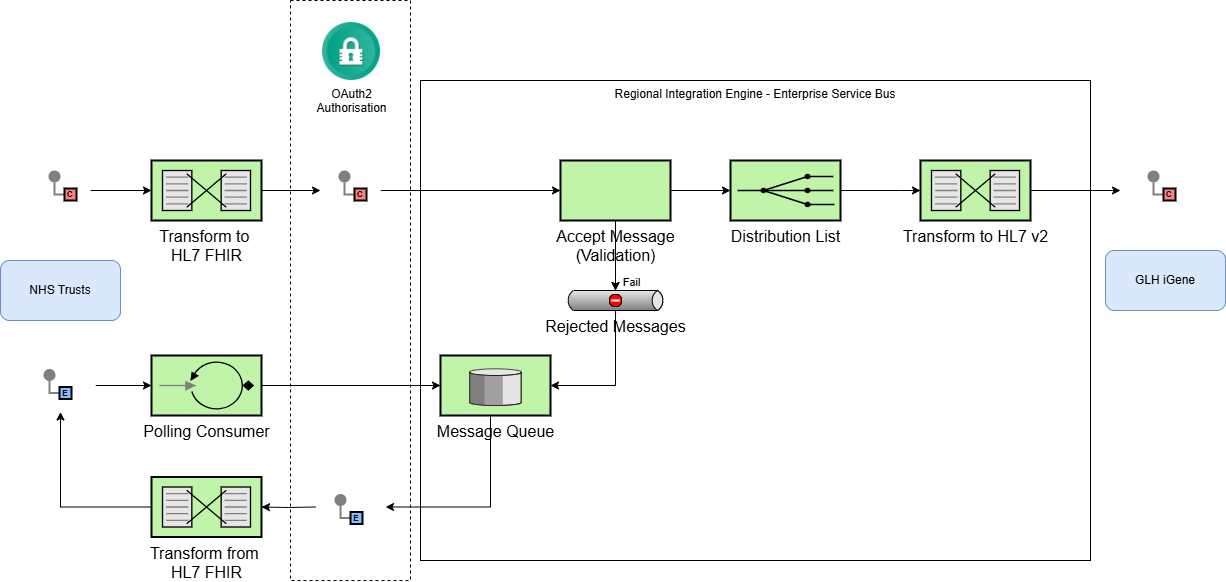
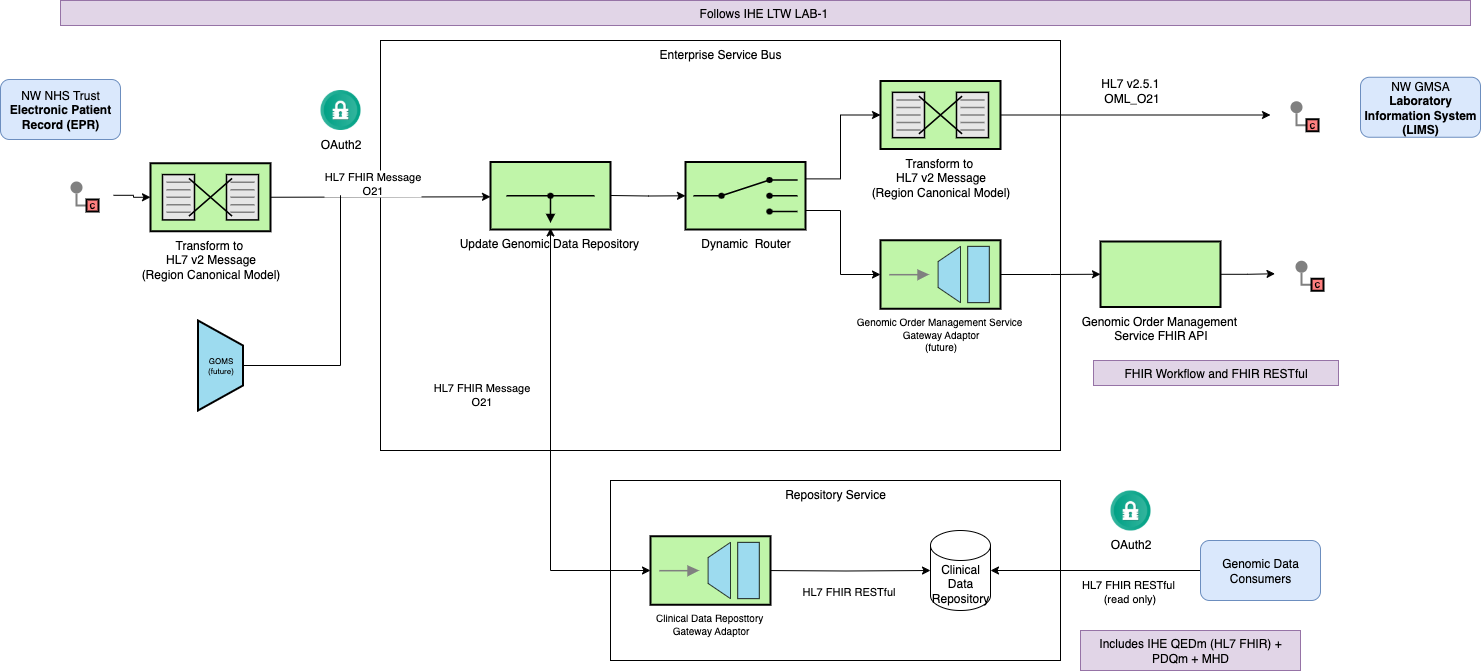
and the North West Regional Integration will provide the adapter from tradtional workflow
to FHIR workflow
|
|
web
|
digital.nhs.uk
|
Future expansions will include notifications from the national Genomic Order Management Service FHIR API
via its FHIR API.
|
|
web
|
www.ihe.net
|
IHE Pathology and Laboratory Medicine (PaLM) Technical Framework - Volume 1
HL7 v2
|
|
web
|
digital.nhs.uk
|
Later stages will include the use of Genomic Order Management Service
.
|
|
web
|
mft.nhs.uk
|
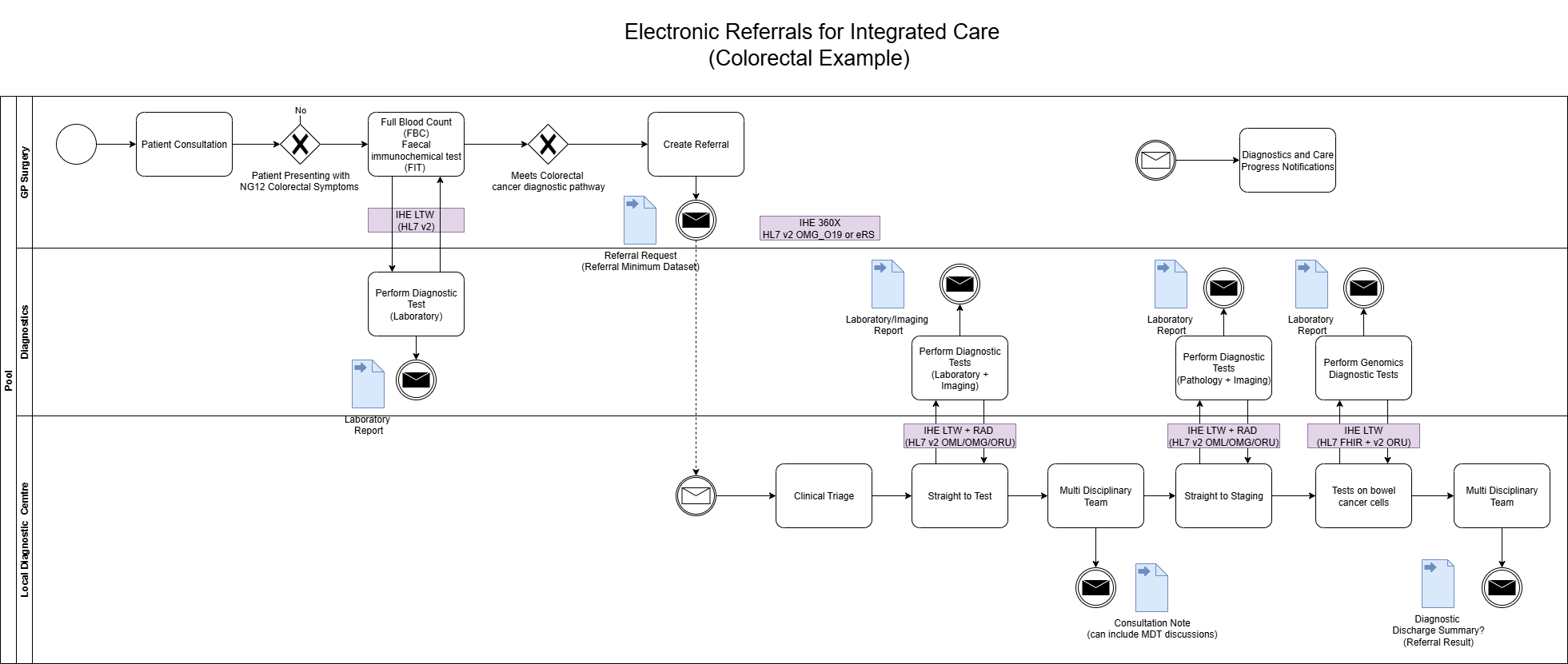
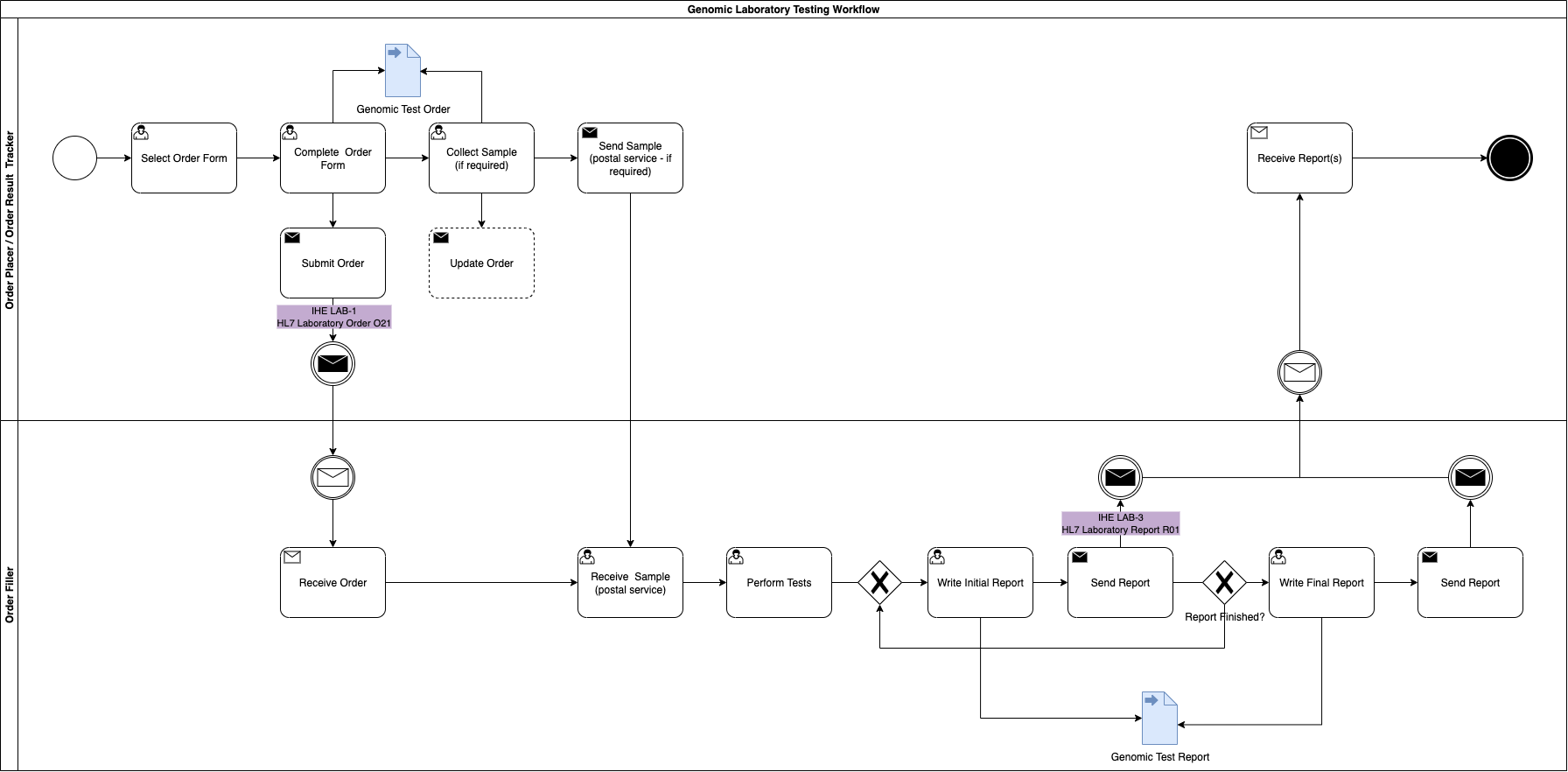
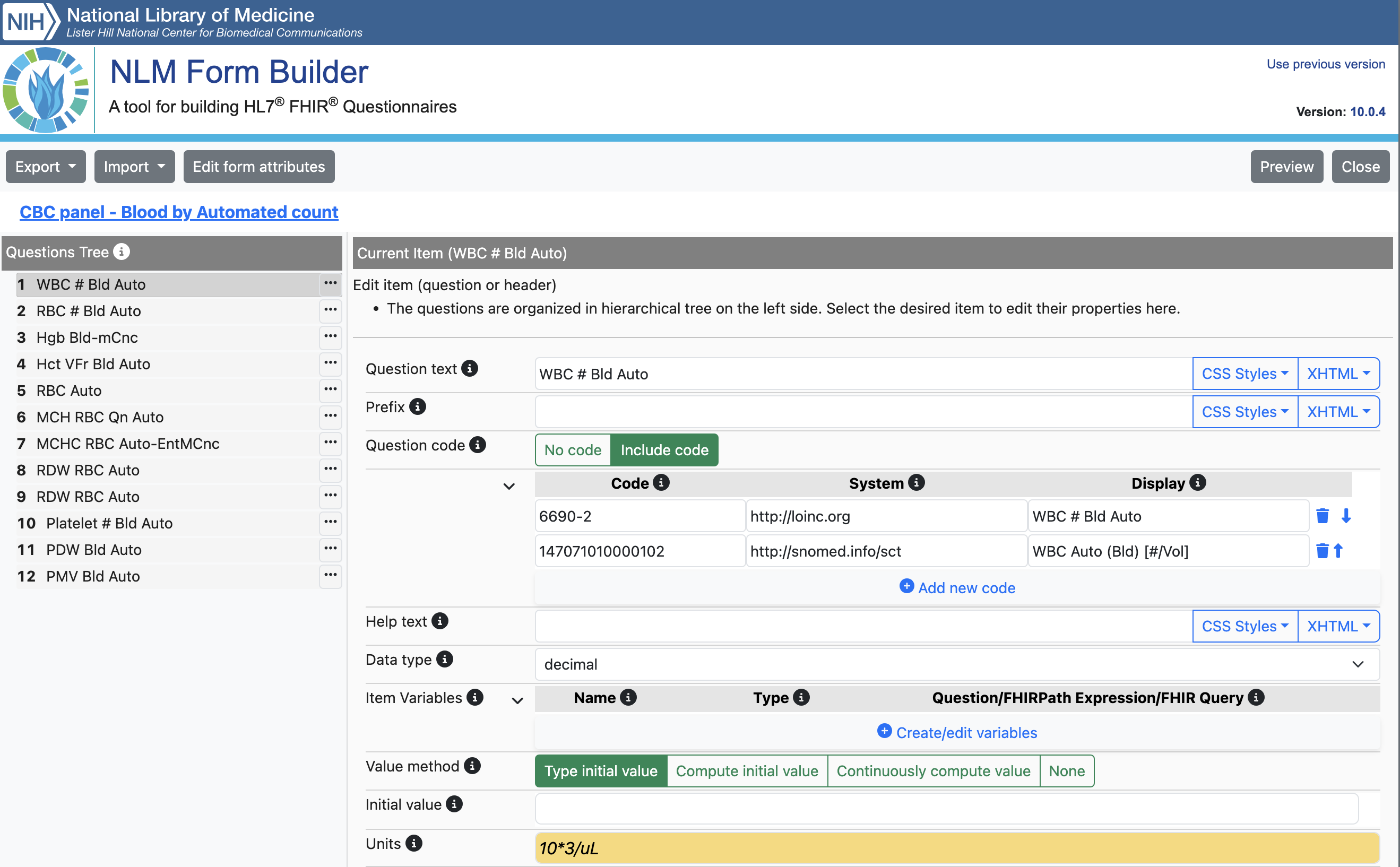
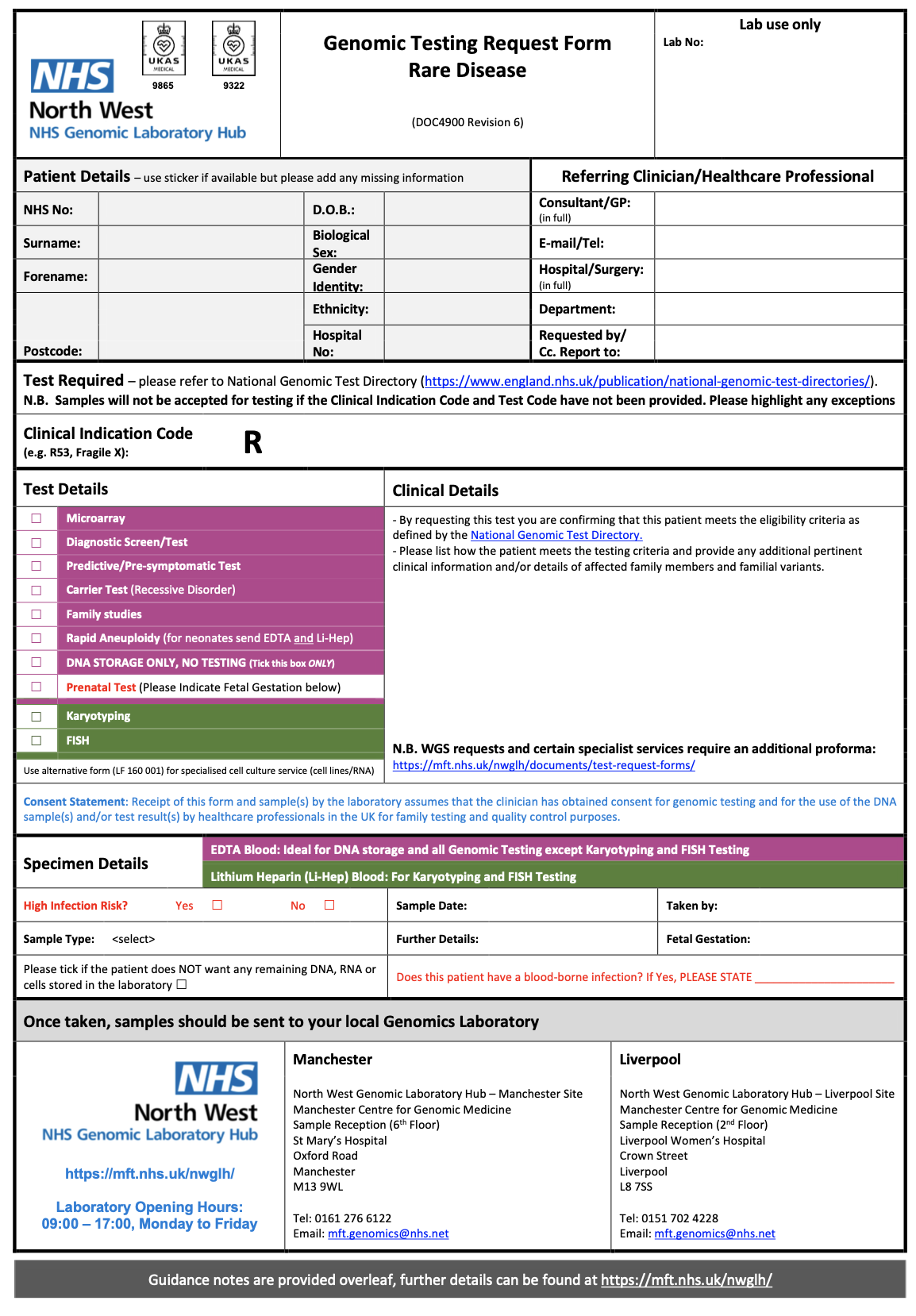
Within the system creating the genomics order, the practitioner will select a form for the test required. Below are several examples from North West Genomic Laboratory Hub - Test Request Forms
.
How this is implemented will vary between different NHS organisations and systems they use.
|
|
web
|
en.wikipedia.org
|
For submission, this form will be converted by the Order Placer
to a communication format called HL7 FHIR
(and for compatability reasons HL7 v2
.
If the Order Placer
has a FHIR enabled Electronic Patient Record (e.g. EPIC, Cerner, Meditech, etc), they may use HL7 SDC - Form Data Extraction
to assist with this process.
|
|
web
|
martinfowler.com
|
This message is an aggregate (DDD)
/ archetype
and so is a collection of FHIR Resources (similar to v2 segements) which is described in Genomic Test Order
.
|
|
web
|
en.wikipedia.org
|
This message is an aggregate (DDD)
/ archetype
and so is a collection of FHIR Resources (similar to v2 segements) which is described in Genomic Test Order
.
|
|
web
|
hl7.eu
|
See also HL7 Europe Laboratory Report - ServiceRequest
This message can be extended by template (FHIR Questionnaire)
which allows the definition of additional questions to be defined for the laboratory order
.
|
|
web
|
profiles.ihe.net
|
It is not expected the NW GLH Laboratory Information Management System (LIMS) will support UK SNOMED CT, and the RIE will handle the conversion either internally using FHIR ConceptMap
or a terminology service with the following capabilities IHE Sharing Valuesets, Codes, and Maps (SVCM)
|
|
web
|
www.ihe.net
|
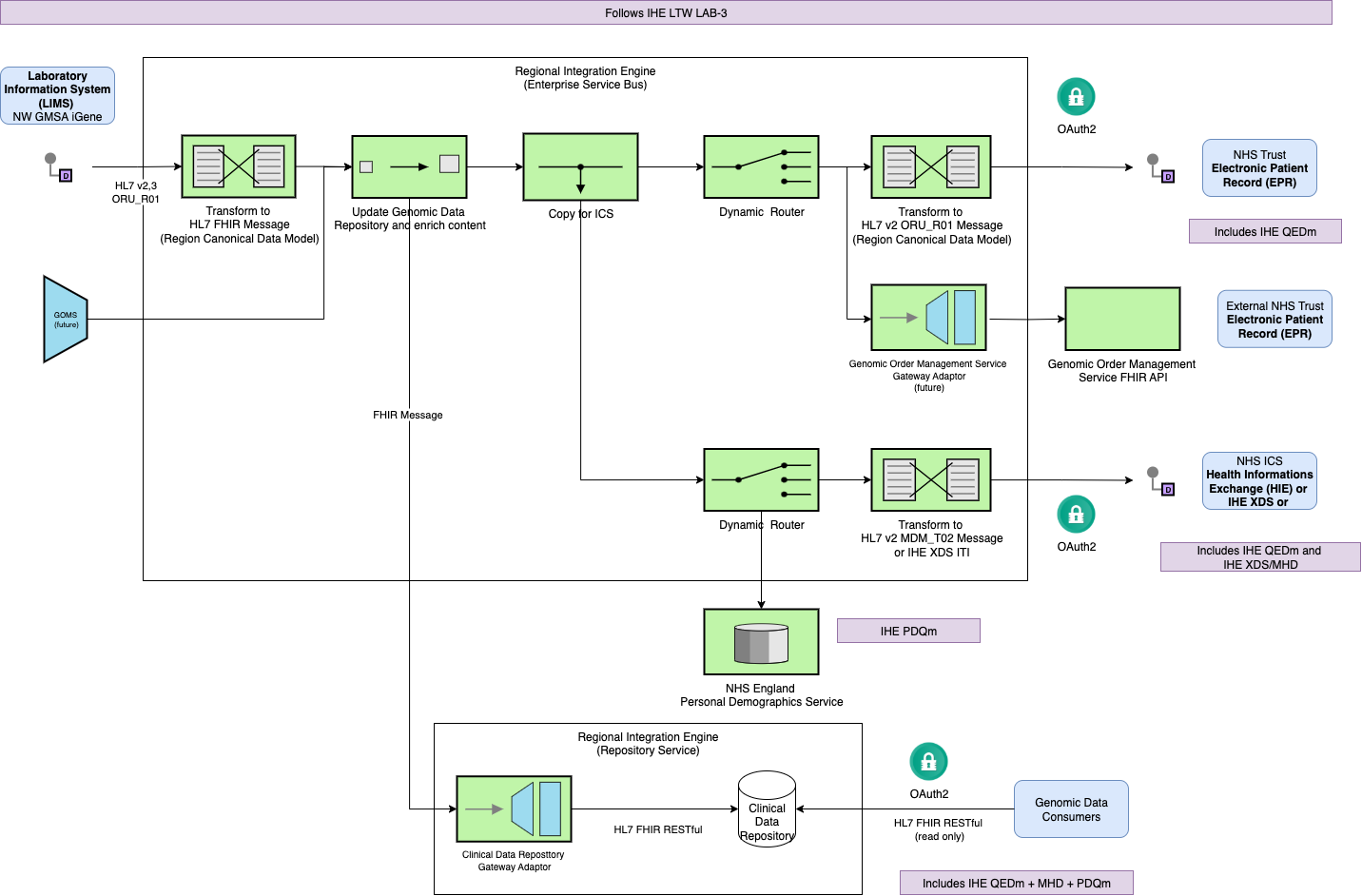
Workflow foundation:
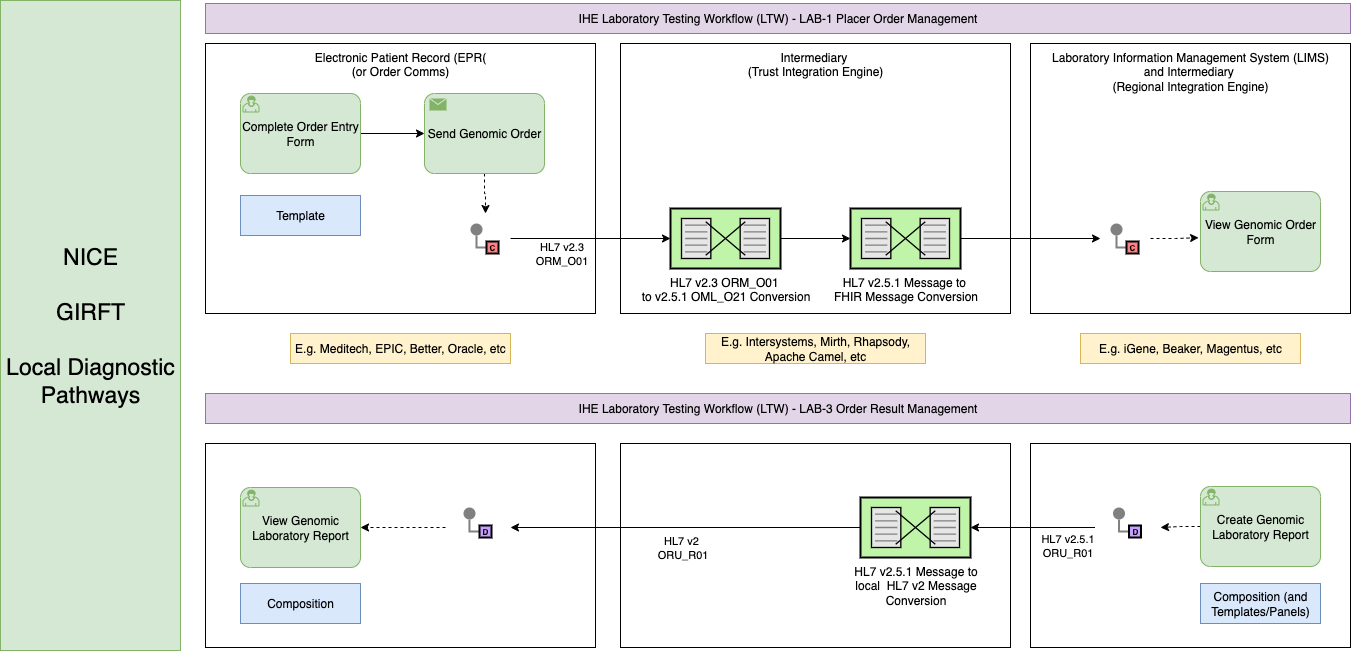
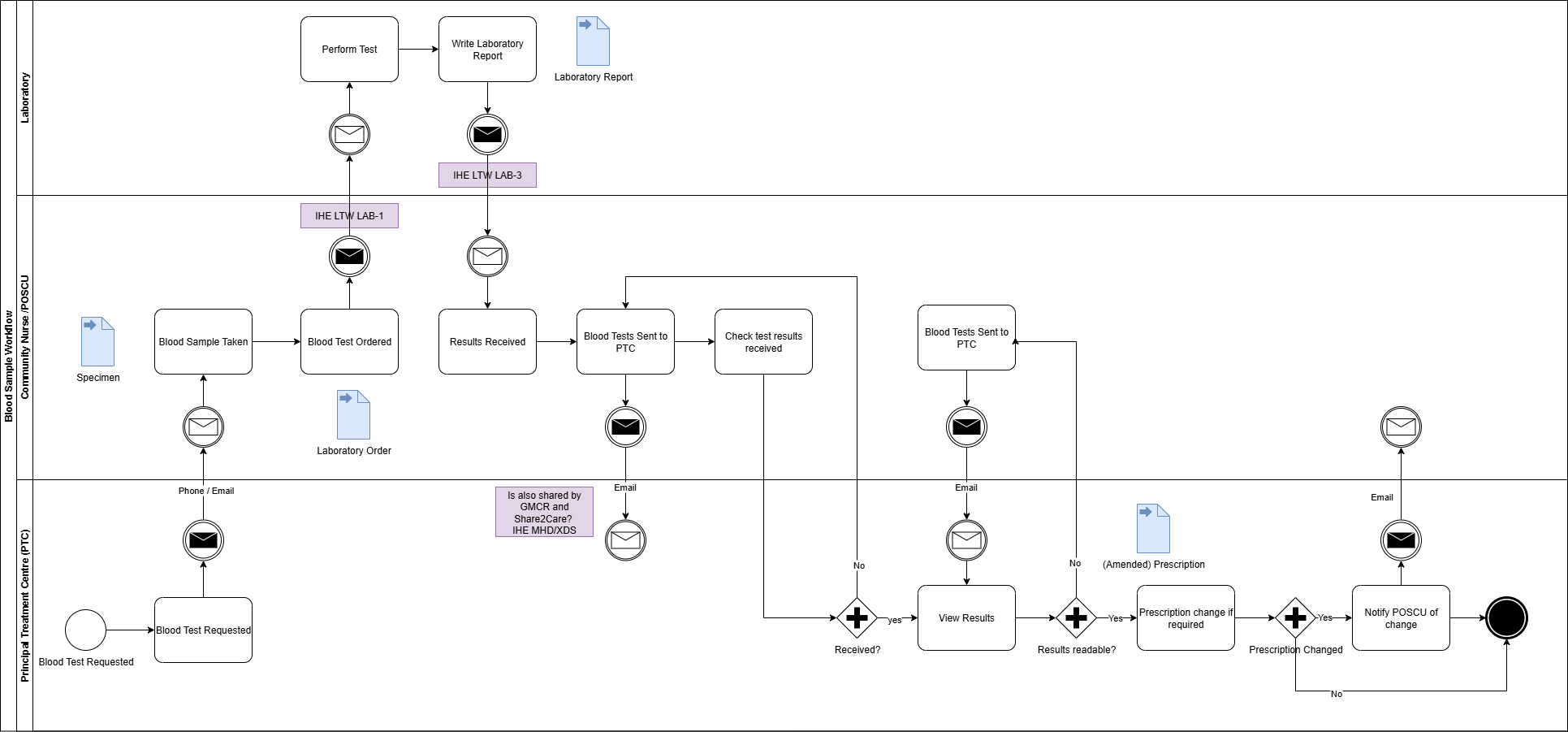
The IHE Laboratory and Testing Worflow LTW
is used as the reference model for describing laboratory testing processes.
|
|
web
|
en.wikipedia.org
|
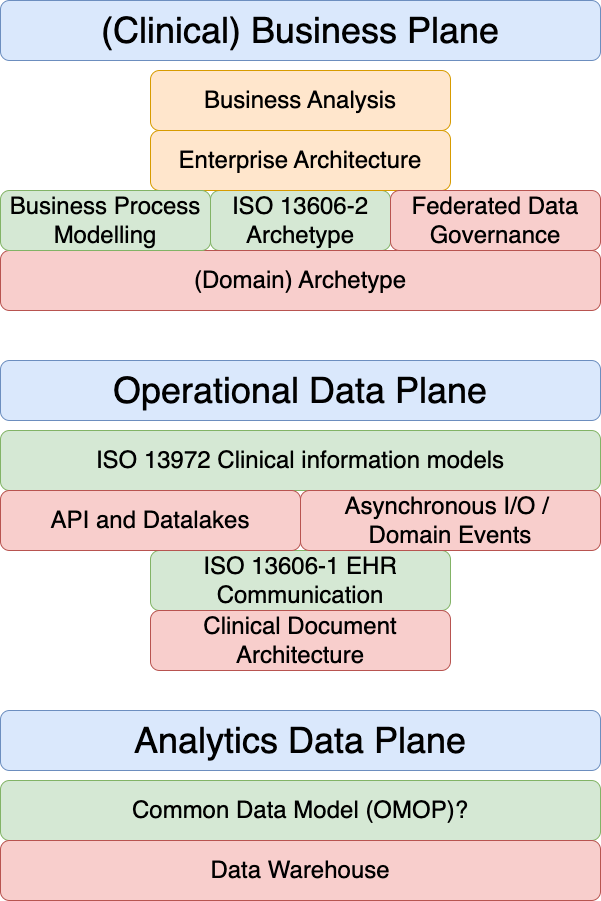
Canonical model:
A standardised model ( Canonical model
), expressed in HL7 FHIR, that can be implemented using HL7 v2, FHIR, and IHE XDS. It aligns with the latest HL7 UK Core and NHS England Data Model and Dictionary. While primarily focused on genomics, it incorporates elements from pathology and radiology for compatibility, and mandates the use of NHS England National Procedure Codes.
|
|
web
|
www.nice.org.uk
|
NICE
standards
|
|
web
|
gettingitrightfirsttime.co.uk
|
Getting It Right First Time (GIRFT)
best practice pathways (e.g. Getting It Right First Time (GIRFT) Best Practice Timed Diagnostic Cancer pathways
|
|
web
|
www.ihe.net
|
These pathways are technically implmented and generally align with the IHE Laboratory and Testing Worflow LTW
(for Imaging workflows see IHE Radiology (RAD)
)
|
|
web
|
www.ihe.net
|
These pathways are technically implmented and generally align with the IHE Laboratory and Testing Worflow LTW
(for Imaging workflows see IHE Radiology (RAD)
)
|
|
web
|
www.ihe.net
|
Workflow basis:
The IHE Laboratory and Testing Worflow LTW
serves as the primary reference for describing laboratory testing processes.
|
|
web
|
wiki.ihe.net
|
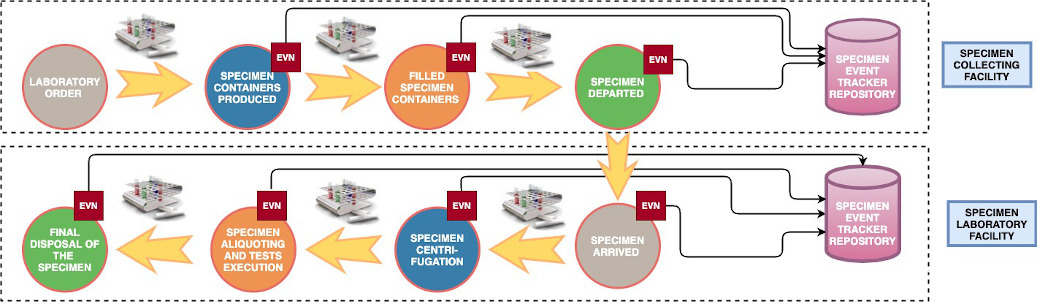
Specimen Event Tracking:
See LAB-40 HL7 v2.9 SET IHE Specimen Event Tracking (SET)
and Hl7 v2.7 OSM_R26 Unsolicited Specimen Shipment Manifest Message
|
|
web
|
hl7-definition.caristix.com
|
Specimen Event Tracking:
See LAB-40 HL7 v2.9 SET IHE Specimen Event Tracking (SET)
and Hl7 v2.7 OSM_R26 Unsolicited Specimen Shipment Manifest Message
|
|
web
|
gettingitrightfirsttime.co.uk
|
The details of this are beyond the scope of this guide, for more details see Getting It Right First Time (GIRFT) Best Practice Timed Diagnostic Cancer pathways
|
|
web
|
www.macmillan.org.uk
|
For information on Genomic Tests on the bowel cancer cells
, see macmillan.org.uk
and NICE DG27 Molecular testing strategies for Lynch syndrome in people with colorectal cancer
|
|
web
|
www.nice.org.uk
|
For information on Genomic Tests on the bowel cancer cells
, see macmillan.org.uk
and NICE DG27 Molecular testing strategies for Lynch syndrome in people with colorectal cancer
|
|
web
|
profiles.ihe.net
|
This may include IHE Internet User Authentication [IUA]
and IHE Basic Audit Log Patterns[BALP]
which includes use of:
|
|
web
|
profiles.ihe.net
|
This may include IHE Internet User Authentication [IUA]
and IHE Basic Audit Log Patterns[BALP]
which includes use of:
|
|
web
|
drive.google.com
|
Note: Event trigger definitions based on NHS England HL7 v2 ADT Message Specification
which is NHS England's supplement to IHE Technical Framework Volume2: Patient Identity Management [ITI-30]
and Patient Encounter Management [ITI-31]
.
|
|
web
|
profiles.ihe.net
|
Note: Event trigger definitions based on NHS England HL7 v2 ADT Message Specification
which is NHS England's supplement to IHE Technical Framework Volume2: Patient Identity Management [ITI-30]
and Patient Encounter Management [ITI-31]
.
|
|
web
|
profiles.ihe.net
|
Note: Event trigger definitions based on NHS England HL7 v2 ADT Message Specification
which is NHS England's supplement to IHE Technical Framework Volume2: Patient Identity Management [ITI-30]
and Patient Encounter Management [ITI-31]
.
|
|
web
|
profiles.ihe.net
|
IHE Patient Administration Management (PAM)
//HL7 v2 ADT Patient Encounter Management (A02, A08 and A12)
|
|
web
|
medcomfhir.dk
|
(Denmark) HL7 FHIR version DK MedCom HospitalNotification
|
|
web
|
profiles.ihe.net
|
IHE Mobile access to Health Documents [MHD]
|
|
web
|
nhsconnect.github.io
|
INTEROPen/NHS England Care Connect API
updated to FHIR R4.
|
|
web
|
profiles.ihe.net
|
Retrieve Document [ITI-68]
|
|
web
|
profiles.ihe.net
|
Find Document References [ITI-67]
|
|
web
|
drive.google.com
|
NHS England HL7 v2 ADT Message Specification
|
|
web
|
drive.google.com
|
NHS England IHE PAM and HL7 v2 Messaging Specification
from NHS England HL7 v2 ADT Message Specification
|
|
web
|
profiles.ihe.net
|
IHE Patient Administration Module (PAM)
HL7 v2
|
|
web
|
profiles.ihe.net
|
Patient Identifier Cross-referencing for mobile (PIXm)
|
|
web
|
profiles.ihe.net
|
Patient Demographics Query for Mobile (PDQm)
|
|
web
|
profiles.ihe.net
|
Patient Master Identity Registry (PMIR)
|
|
web
|
profiles.ihe.net
|
Patient Identity Feed (ITI-8)
Patient Identity Feed FHIR (ITI-104)
Mobile Patient Identity Feed (ITI-93)
|
|
web
|
profiles.ihe.net
|
Patient Identity Feed (ITI-8)
Patient Identity Feed FHIR (ITI-104)
Mobile Patient Identity Feed (ITI-93)
|
|
web
|
profiles.ihe.net
|
Patient Identity Feed (ITI-8)
Patient Identity Feed FHIR (ITI-104)
Mobile Patient Identity Feed (ITI-93)
|
|
web
|
profiles.ihe.net
|
The Patient Identity Source
sends a IHE ITI-8/HL7 v2 A31 (or ITI-104
/FHIR PUT (upsert)) event to the RIE
|
|
web
|
digital.nhs.uk
|
Lookup up existing Patient via supplied identifiers (MRN or NHS Number) via a FHIR RESTful Search
.
|
|
web
|
digital.nhs.uk
|
If a Patient record is found, then update the Patient record via a FHIR RESTful Update
|
|
web
|
profiles.ihe.net
|
Mobile Patient Demographics Query (ITI-78)
Patient Demographics Query (ITI-21)
|
|
web
|
profiles.ihe.net
|
Mobile Patient Demographics Query (ITI-78)
Patient Demographics Query (ITI-21)
|
|
web
|
profiles.ihe.net
|
IHE Mobile Patient Demographics Query [ITI-78]
|
|
web
|
fhir.interweavedigital.com
|
This Encounter represents the overall episode or stay, for further information see Interweave Encounter Grouping
|
|
web
|
project-wildfyre.github.io
|
Questionnaire Viewer
|
|
web
|
project-wildfyre.github.io
|
Questionnaire Viewer
|
|
web
|
project-wildfyre.github.io
|
Master HL7 genetic variant reporting panel Questionnaire Viewer
|
|
web
|
project-wildfyre.github.io
|
Questionnaire Viewer
|
|
web
|
www.england.nhs.uk
|
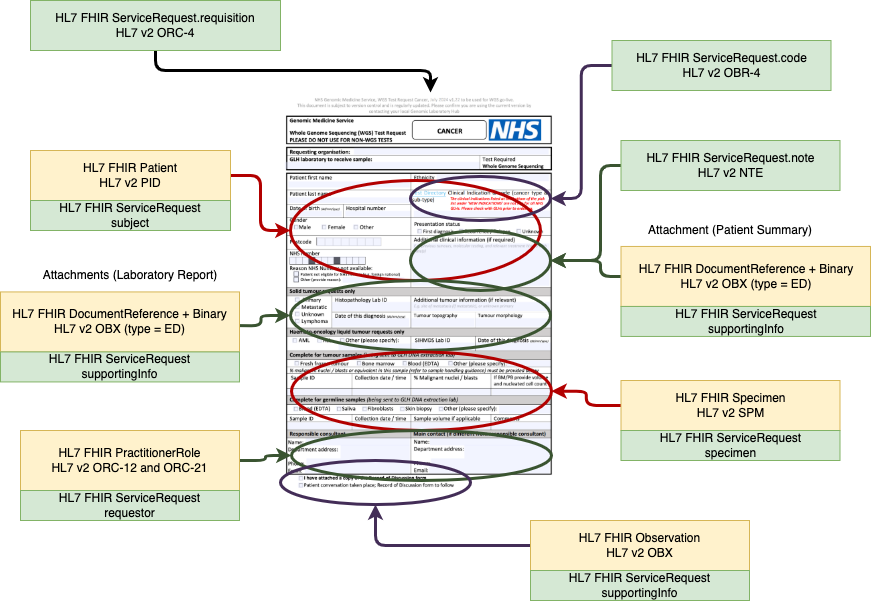
NHS Genomic Medicine Service test order forms
|
|
web
|
project-wildfyre.github.io
|
Archetype Viewer Questionnaire-GenomicTestOrder
|
|
web
|
www.datadictionary.nhs.uk
|
Genomic Laboratory Order
Diagnostic Order
Patient Administration
Patient
Identification
NHSNumber
Identification
Medical Record Number
Name, date of birth, ethnic category, etc.
Case identification / Account Number
Hospital Provider Spell Identifier
Laboratory Order
Order Details
Placer Order Number
Filler Order Number
Placer Group Number
Ordering Facility
Organisation Code
Ordering Practitioner
Gene disease assessed
Clinical Indication
Local Treatment Category Code
Genomic Test Code
Test Directory Code
Notes
Specimen
Specimen Id
Accession Number
Shipment Tracking number
Order Entry Questions
Genomic
(Supporting Information)
Consent
Pregnant
Consanguineous union
|
|
web
|
nw-gmsa.github.io
|
Patient
|
|
web
|
ckm.openehr.org
|
C. openEHR Genomics Project
|
|
web
|
project-wildfyre.github.io
|
Archetype Viewer Questionnaire-GenomicTestReport
|
|
web
|
www.datadictionary.nhs.uk
|
Genomic Laboratory Report
Diagnostic Report
Patient Administration
Patient
Identification
NHSNumber
Identification
Medical Record Number
Name, date of birth, ethnic category, etc.
Case identification
Hospital Provider Spell Identifier
Laboratory Report
Order Details
Placer Order Number
Filler Order Number
Ordering Facility
Organisation Code
Gene disease assessed
Clinical Indication
Local Treatment Category Code
Genomic Test Code
Test Directory Code
Specimen
Accession Number
Test Details
Report Number
Performer
Results Interpreter
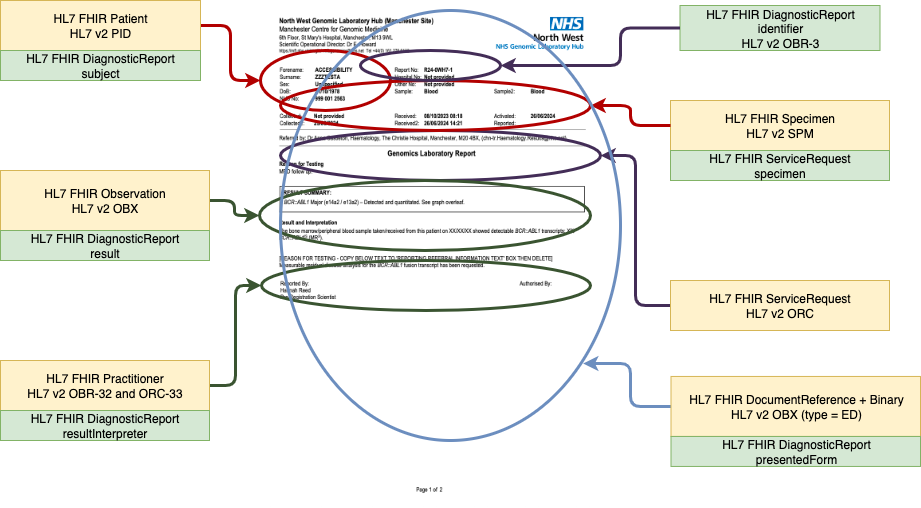
Conclusion (Clinical Indication)
Results Genomic
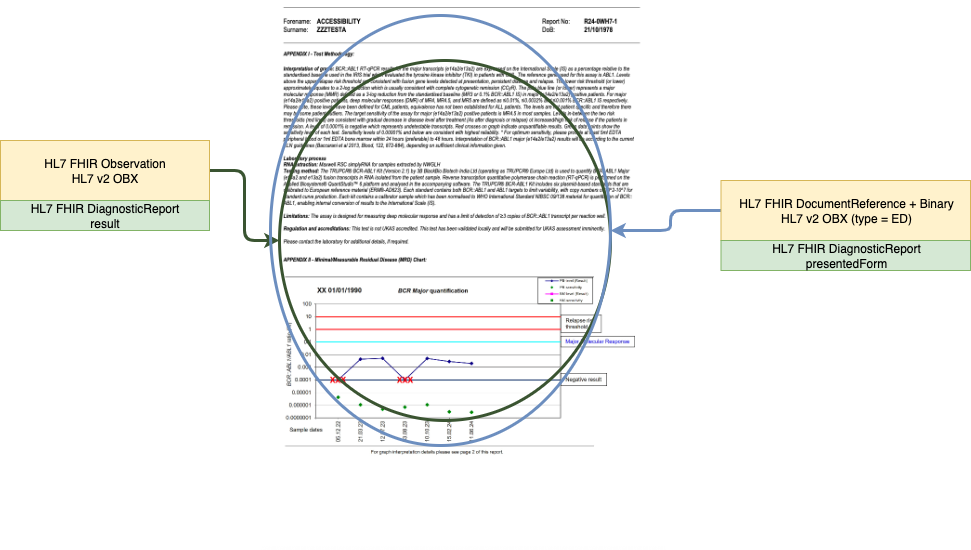
Genomic Analysis Narrative Report (PDF)
Attachment
Finding
Variant
Implication
Diagnostic Implication
Therapeutic Implication
ProgressRX
Genomic Study/Method
Gene disease assessed / Clinical Indication
Analysis method [Type]
NHS England Genomics Test Outcome
Genomic Study Analysis
Regions Studied / Gene mutations tested
|
|
web
|
nw-gmsa.github.io
|
Placer Order Number
|
|
web
|
pedigree.readthedocs.io
|
GA4GH Pedigree - Conceptual Model
|
|
web
|
mft.nhs.uk
|
NW Genomics - Gen O
|
|
web
|
project-wildfyre.github.io
|
Archetype Viewer Questionnaire-Pedigree
|
|
web
|
wiki.ihe.net
|
IHE Specimen Event Tracking
|
|
web
|
mft.nhs.uk
|
NHS MFT Transport of Samples
|
|
web
|
profiles.ihe.net
|
Core model following IHE Basic Audit Log Patterns (BALP)
|
|
web
|
www.ehealth.fgov.be
|
(copied from BE eHealth Platform Federal Core Profiles
) Extension able to hold a reference and a concept (Temporary solution until https://jira.hl7.org/browse/FHIR-44661 is solved and see Zulip: https://chat.fhir.org/#narrow/stream/179280-fhir.2Finfrastructure-wg/topic/Backporting.20CodeableReference )
|
|
web
|
wiki.ihe.net
|
This is a metadata field from XDS/MHD
.
|
|
web
|
ckm.openehr.org
|
openEHR Project: Genomics
|
|
web
|
www.england.nhs.uk
|
NHS England Genomics Test Reporting Specification
|
|
web
|
drive.google.com
|
NHS England HL7 v2
DG1 ADT Message Specification
|
|
web
|
www.archi-lab.io
|
Rule 3: Reference Other Aggregates by Identity (Implementing Domain Driven Design)
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Enterprise Integration Patterns - Correlation Identifier
.
|
|
web
|
hl7-definition.caristix.com
|
HL7 v2 EI - Entity Identifier
|
|
web
|
hl7-definition.caristix.com
|
HL7 v2 CX - Extended Composite ID with Check Digit
|
|
web
|
www.rcr.ac.uk
|
The Royal College of Radiologists Reporting networks - understanding the technical options
|
|
web
|
www.ihe-europe.net
|
Diagnostic
Reference: IHE Europe Metadata for exchange medical documents and images
|
|
web
|
www.ihe-europe.net
|
IHE Europe Document Metadata
(this contains references to NHS England terminology)
|
|
web
|
www.digihealthcare.scot
|
Digital Health and Care Scotland - (EH4001) CLINICAL DOCUMENT INDEXING STANDARDS
|
|
web
|
profiles.ihe.net
|
Note: links to HL7 FHIR Practitioner has removed several IHE XDS Document Entry fields, and just retained identifier and name. It is expected these values would be available via the NHS England Healthcare Worker API which may conform to IHE Mobile Care Services Discovery (mCSD)
interface contracts.
|
|
web
|
www.datadictionary.nhs.uk
|
ACTIVITY LOCATION TYPE CODE
|
|
web
|
drive.google.com
|
NHS England HL7 v2
PV1 ADT Message Specification
|
|
web
|
fhir.interweavedigital.com
|
For detailed notes on FHIR Encounter in a NHS region, see Yorkshire and Humberside Care Record (YHCR) - FHIR Encounter
( YHCR GitHub Repository
)
|
|
web
|
github.com
|
For detailed notes on FHIR Encounter in a NHS region, see Yorkshire and Humberside Care Record (YHCR) - FHIR Encounter
( YHCR GitHub Repository
)
|
|
web
|
digital.nhs.uk
|
NHS England - Multicast Notification Service API
|
|
web
|
www.datadictionary.nhs.uk
|
Patient Administration
'NHS Data Model and Dictionary' HOSPITAL PROVIDER SPELL IDENTIFIER
|
|
web
|
drive.google.com
|
HL7 v2
Visit Number (PV1-19). See also NHS England HL7 v2 ADT Message Specification
PV1 documentation.
|
|
web
|
fhir.interweavedigital.com
|
Patient Administration
Related to Yorkskhire and Humberside - EncounterGrouping
|
|
web
|
www.datadictionary.nhs.uk
|
Patient Administration
'NHS Data Model and Dictionary' LOCAL PATIENT IDENTIFIER
|
|
web
|
www.datadictionary.nhs.uk
|
NHS Data Model and Dictionary
England/Wales NHS NUMBER
, Northern Ireland HEALTH AND CARE NUMBER
and Scotland COMMUNITY HEALTH INDEX NUMBER
Only traced NHS Number SHOULD
be used, un-traced NHS Numbers MUST
be clearly indicated.
|
|
web
|
www.datadictionary.nhs.uk
|
NHS Data Model and Dictionary
England/Wales NHS NUMBER
, Northern Ireland HEALTH AND CARE NUMBER
and Scotland COMMUNITY HEALTH INDEX NUMBER
Only traced NHS Number SHOULD
be used, un-traced NHS Numbers MUST
be clearly indicated.
|
|
web
|
www.datadictionary.nhs.uk
|
NHS Data Model and Dictionary
England/Wales NHS NUMBER
, Northern Ireland HEALTH AND CARE NUMBER
and Scotland COMMUNITY HEALTH INDEX NUMBER
Only traced NHS Number SHOULD
be used, un-traced NHS Numbers MUST
be clearly indicated.
|
|
web
|
www.datadictionary.nhs.uk
|
Format: NNNNNNNNNN, values include a checksum, details can be found in NHS NUMBER
|
|
web
|
en.wikipedia.org
|
Secondary findings are genetic test results that provide information about variants in a gene unrelated to the primary purpose for the testing, most often discovered when Whole Exome Sequencing (WES)
or Whole Genome Sequencing (WGS)
is performed. This extension should be used to denote when a genetic finding is being shared as a secondary finding, and ideally refer to a corresponding guideline or policy statement.
|
|
web
|
en.wikipedia.org
|
Secondary findings are genetic test results that provide information about variants in a gene unrelated to the primary purpose for the testing, most often discovered when Whole Exome Sequencing (WES)
or Whole Genome Sequencing (WGS)
is performed. This extension should be used to denote when a genetic finding is being shared as a secondary finding, and ideally refer to a corresponding guideline or policy statement.
|
|
web
|
drive.google.com
|
NHS England HL7 v2
OBX ADT Message Specification
|
|
web
|
www.datadictionary.nhs.uk
|
Patient Administration
'NHS Data Model and Dictionary' ORGANISATION_CODE
|
|
web
|
www.datadictionary.nhs.uk
|
Care Directory
'NHS Data Model and Dictionary' ORGANISATION SITE IDENTIFIER
|
|
web
|
drive.google.com
|
NHS England HL7 v2
XON ADT Message Specification
|
|
web
|
hl7-definition.caristix.com
|
HL7 v2 XON – Extended Composite Name and Identification Number for Organisations
|
|
web
|
profiles.ihe.net
|
IHE Mobile Care Services Discovery (mCSD)
|
|
web
|
digital.nhs.uk
|
This reference
may be able to point to Care Service Directory
services provided by NHS England Organisation Data Terminology - FHIR API
(or alternatively downloaded from NHS England Organisation Data Service - CSV Downloads
, therefore it is proposed
|
|
web
|
digital.nhs.uk
|
This reference
may be able to point to Care Service Directory
services provided by NHS England Organisation Data Terminology - FHIR API
(or alternatively downloaded from NHS England Organisation Data Service - CSV Downloads
, therefore it is proposed
|
|
web
|
drive.google.com
|
NHS England HL7 v2
PID ADT Message Specification
|
|
web
|
drive.google.com
|
NHS England HL7 v2
XCN ADT Message Specification
|
|
web
|
hl7-definition.caristix.com
|
HL7 v2 XCN Extended Composite ID Number and Name for Persons
|
|
web
|
www.datadictionary.nhs.uk
|
It is not clear in Enterprise/Regional use of FHIR which approach Resource
or Reference
should be taken, both HL7 v2 and FHIR support Reference aggregate or entities by identity
from Dommain Driven Design (DDD) and this appears to also be followed by IHE XDS and DICOM.
In addition, NHS England Data Dictionary
and NHS England HL7 v2 ADT Message Specification
favour Reference
NHS England FHIR STU3/R4 specifications around Messaging, tend to favour Resource
.
|
|
web
|
digital.nhs.uk
|
It is likely that the reference
may be able to point to Care Service Directory
services provided by NHS England Healthcare Worker - FHIR API
in the near future,(or alternatively downloaded from NHS England Organisation Data Service - CSV Downloads
. Therefore: it is proposed:
|
|
web
|
digital.nhs.uk
|
It is likely that the reference
may be able to point to Care Service Directory
services provided by NHS England Healthcare Worker - FHIR API
in the near future,(or alternatively downloaded from NHS England Organisation Data Service - CSV Downloads
. Therefore: it is proposed:
|
|
web
|
www.datadictionary.nhs.uk
|
'Care Directory' 'NHS Data Model and Dictionary' CONSULTANT CODE
and GENERAL MEDICAL PRACTITIONER PPD CODE
.
|
|
web
|
www.datadictionary.nhs.uk
|
'Care Directory' 'NHS Data Model and Dictionary' CONSULTANT CODE
and GENERAL MEDICAL PRACTITIONER PPD CODE
.
|
|
web
|
www.datadictionary.nhs.uk
|
Uniquely identifies a Specimen across multiple laboratory systems.
This is related to RADIOLOGICAL ACCESSION NUMBER
which applies to Imaging Studies.
|
|
web
|
www.ihe.net
|
IHE Pathology and Laboratory Medicine (PaLM) Volume 2 - LAB-4
from the LIMS, offers a potential approach to handling this.
|
|
web
|
www.ihe.net
|
IHE Pathology and Laboratory Medicine (PaLM) Volume 2 - LAB-4
from the LIMS, which can be issued whenever specimen details are updated.
|
|
web
|
fhir.interweavedigital.com
|
Patient Administration
Equivalent to Yorkskhire and Humberside - Encounter
|
|
web
|
www.ehealth.fgov.be
|
Simple Extension with the type CodeableConcept: (copied from BE eHealth Platform Federal Core Profiles
) Extension able to hold a reference and a concept (Temporary solution until https://jira.hl7.org/browse/FHIR-44661 is solved and see Zulip: https://chat.fhir.org/#narrow/stream/179280-fhir.2Finfrastructure-wg/topic/Backporting.20CodeableReference )
|
|
web
|
www.england.nhs.uk
|
See National genomic test directory for cancer
on Genomic Test Directory
|
|
web
|
snomed.info
|
47461006
|
|
web
|
snomed.info
|
66511000119102
|
|
web
|
snomed.info
|
66551000119101
|
|
web
|
snomed.info
|
66601000119104
|
|
web
|
snomed.info
|
66621000119108
|
|
web
|
snomed.info
|
66631000119106
|
|
web
|
snomed.info
|
66661000119103
|
|
web
|
snomed.info
|
66721000119101
|
|
web
|
snomed.info
|
66731000119103
|
|
web
|
snomed.info
|
66751000119109
|
|
web
|
snomed.info
|
66781000119102
|
|
web
|
snomed.info
|
66971000119103
|
|
web
|
snomed.info
|
90671000119109
|
|
web
|
snomed.info
|
98311000119105
|
|
web
|
snomed.info
|
98421000119108
|
|
web
|
snomed.info
|
137511000119103
|
|
web
|
snomed.info
|
286481000119102
|
|
web
|
snomed.info
|
302571000000101
|
|
web
|
snomed.info
|
384511000000103
|
|
web
|
snomed.info
|
384671000000109
|
|
web
|
snomed.info
|
750571000000106
|
|
web
|
snomed.info
|
750601000000104
|
|
web
|
snomed.info
|
1128411000000105
|
|
web
|
snomed.info
|
1219071000000109
|
|
web
|
snomed.info
|
1219081000000106
|
|
web
|
snomed.info
|
1219171000000108
|
|
web
|
snomed.info
|
1219181000000105
|
|
web
|
snomed.info
|
1219301000000103
|
|
web
|
snomed.info
|
481462461000119102
|
|
web
|
snomed.info
|
559151051000119108
|
|
web
|
snomed.info
|
680035451000119103
|
|
web
|
snomed.info
|
697788021000119105
|
|
web
|
snomed.info
|
106221001
|
|
web
|
snomed.info
|
816009
|
|
web
|
snomed.info
|
842009
|
|
web
|
snomed.info
|
1168007
|
|
web
|
snomed.info
|
1318006
|
|
web
|
snomed.info
|
2308003
|
|
web
|
snomed.info
|
2351004
|
|
web
|
snomed.info
|
3067005
|
|
web
|
snomed.info
|
6800004
|
|
web
|
snomed.info
|
7599007
|
|
web
|
snomed.info
|
8116006
|
|
web
|
snomed.info
|
9109004
|
|
web
|
snomed.info
|
10589004
|
|
web
|
snomed.info
|
12645001
|
|
web
|
snomed.info
|
13300001
|
|
web
|
snomed.info
|
13333006
|
|
web
|
snomed.info
|
14915001
|
|
web
|
snomed.info
|
16345006
|
|
web
|
snomed.info
|
17523003
|
|
web
|
snomed.info
|
19482002
|
|
web
|
snomed.info
|
22061001
|
|
web
|
snomed.info
|
22986007
|
|
web
|
snomed.info
|
24403008
|
|
web
|
snomed.info
|
25132006
|
|
web
|
snomed.info
|
25194005
|
|
web
|
snomed.info
|
25363001
|
|
web
|
snomed.info
|
25384006
|
|
web
|
snomed.info
|
25900007
|
|
web
|
snomed.info
|
26608005
|
|
web
|
snomed.info
|
28820008
|
|
web
|
snomed.info
|
29286002
|
|
web
|
snomed.info
|
29549004
|
|
web
|
snomed.info
|
31091003
|
|
web
|
snomed.info
|
32475006
|
|
web
|
snomed.info
|
34216002
|
|
web
|
snomed.info
|
34782005
|
|
web
|
snomed.info
|
34850003
|
|
web
|
snomed.info
|
35147005
|
|
web
|
snomed.info
|
37819008
|
|
web
|
snomed.info
|
38194003
|
|
web
|
snomed.info
|
38789009
|
|
web
|
snomed.info
|
39751009
|
|
web
|
snomed.info
|
40976007
|
|
web
|
snomed.info
|
41482005
|
|
web
|
snomed.info
|
43245005
|
|
web
|
snomed.info
|
43376001
|
|
web
|
snomed.info
|
45212007
|
|
web
|
snomed.info
|
45427005
|
|
web
|
snomed.info
|
45597001
|
|
web
|
snomed.info
|
45803000
|
|
web
|
snomed.info
|
47708004
|
|
web
|
snomed.info
|
47986005
|
|
web
|
snomed.info
|
50334000
|
|
web
|
snomed.info
|
50606000
|
|
web
|
snomed.info
|
51512005
|
|
web
|
snomed.info
|
52816004
|
|
web
|
snomed.info
|
54236009
|
|
web
|
snomed.info
|
54828006
|
|
web
|
snomed.info
|
55446002
|
|
web
|
snomed.info
|
56879003
|
|
web
|
snomed.info
|
57196006
|
|
web
|
snomed.info
|
57652005
|
|
web
|
snomed.info
|
58460004
|
|
web
|
snomed.info
|
60181007
|
|
web
|
snomed.info
|
60844005
|
|
web
|
snomed.info
|
61668005
|
|
web
|
snomed.info
|
61828008
|
|
web
|
snomed.info
|
63693001
|
|
web
|
snomed.info
|
64195000
|
|
web
|
snomed.info
|
64245008
|
|
web
|
snomed.info
|
64553001
|
|
web
|
snomed.info
|
65087006
|
|
web
|
snomed.info
|
65247007
|
|
web
|
snomed.info
|
69604007
|
|
web
|
snomed.info
|
69868005
|
|
web
|
snomed.info
|
72633008
|
|
web
|
snomed.info
|
72644000
|
|
web
|
snomed.info
|
73804003
|
|
web
|
snomed.info
|
74354009
|
|
web
|
snomed.info
|
74428002
|
|
web
|
snomed.info
|
74836001
|
|
web
|
snomed.info
|
78473004
|
|
web
|
snomed.info
|
79248008
|
|
web
|
snomed.info
|
79667004
|
|
web
|
snomed.info
|
81835007
|
|
web
|
snomed.info
|
81919004
|
|
web
|
snomed.info
|
82283002
|
|
web
|
snomed.info
|
83579008
|
|
web
|
snomed.info
|
85559002
|
|
web
|
snomed.info
|
85599004
|
|
web
|
snomed.info
|
85900004
|
|
web
|
snomed.info
|
85938000
|
|
web
|
snomed.info
|
86457007
|
|
web
|
snomed.info
|
87682005
|
|
web
|
snomed.info
|
88942003
|
|
web
|
snomed.info
|
89017001
|
|
web
|
snomed.info
|
89109006
|
|
web
|
snomed.info
|
89551006
|
|
web
|
snomed.info
|
89744004
|
|
web
|
snomed.info
|
103225004
|
|
web
|
snomed.info
|
110440006
|
|
web
|
snomed.info
|
110441005
|
|
web
|
snomed.info
|
110442003
|
|
web
|
snomed.info
|
112143006
|
|
web
|
snomed.info
|
112144000
|
|
web
|
snomed.info
|
112149005
|
|
web
|
snomed.info
|
112211004
|
|
web
|
snomed.info
|
112212006
|
|
web
|
snomed.info
|
115665000
|
|
web
|
snomed.info
|
115730009
|
|
web
|
snomed.info
|
115731008
|
|
web
|
snomed.info
|
115732001
|
|
web
|
snomed.info
|
115734000
|
|
web
|
snomed.info
|
115735004
|
|
web
|
snomed.info
|
115736003
|
|
web
|
snomed.info
|
115737007
|
|
web
|
snomed.info
|
115748000
|
|
web
|
snomed.info
|
115749008
|
|
web
|
snomed.info
|
115750008
|
|
web
|
snomed.info
|
115751007
|
|
web
|
snomed.info
|
115752000
|
|
web
|
snomed.info
|
115753005
|
|
web
|
snomed.info
|
115754004
|
|
web
|
snomed.info
|
115755003
|
|
web
|
snomed.info
|
115756002
|
|
web
|
snomed.info
|
115758001
|
|
web
|
snomed.info
|
115759009
|
|
web
|
snomed.info
|
115760004
|
|
web
|
snomed.info
|
115761000
|
|
web
|
snomed.info
|
115762007
|
|
web
|
snomed.info
|
115763002
|
|
web
|
snomed.info
|
115764008
|
|
web
|
snomed.info
|
115794002
|
|
web
|
snomed.info
|
115795001
|
|
web
|
snomed.info
|
115796000
|
|
web
|
snomed.info
|
115797009
|
|
web
|
snomed.info
|
115798004
|
|
web
|
snomed.info
|
115799007
|
|
web
|
snomed.info
|
115800006
|
|
web
|
snomed.info
|
115801005
|
|
web
|
snomed.info
|
115802003
|
|
web
|
snomed.info
|
115803008
|
|
web
|
snomed.info
|
115804002
|
|
web
|
snomed.info
|
115805001
|
|
web
|
snomed.info
|
115821006
|
|
web
|
snomed.info
|
115822004
|
|
web
|
snomed.info
|
115823009
|
|
web
|
snomed.info
|
115824003
|
|
web
|
snomed.info
|
115825002
|
|
web
|
snomed.info
|
115826001
|
|
web
|
snomed.info
|
115827005
|
|
web
|
snomed.info
|
115830003
|
|
web
|
snomed.info
|
115831004
|
|
web
|
snomed.info
|
115832006
|
|
web
|
snomed.info
|
115833001
|
|
web
|
snomed.info
|
115834007
|
|
web
|
snomed.info
|
115835008
|
|
web
|
snomed.info
|
115837000
|
|
web
|
snomed.info
|
115838005
|
|
web
|
snomed.info
|
115839002
|
|
web
|
snomed.info
|
115844009
|
|
web
|
snomed.info
|
115845005
|
|
web
|
snomed.info
|
115851000
|
|
web
|
snomed.info
|
115852007
|
|
web
|
snomed.info
|
115853002
|
|
web
|
snomed.info
|
115854008
|
|
web
|
snomed.info
|
115855009
|
|
web
|
snomed.info
|
115860008
|
|
web
|
snomed.info
|
115861007
|
|
web
|
snomed.info
|
115866002
|
|
web
|
snomed.info
|
115867006
|
|
web
|
snomed.info
|
115940004
|
|
web
|
snomed.info
|
118205009
|
|
web
|
snomed.info
|
124972006
|
|
web
|
snomed.info
|
124975008
|
|
web
|
snomed.info
|
124976009
|
|
web
|
snomed.info
|
124977000
|
|
web
|
snomed.info
|
124979002
|
|
web
|
snomed.info
|
124980004
|
|
web
|
snomed.info
|
124981000
|
|
web
|
snomed.info
|
124982007
|
|
web
|
snomed.info
|
124988006
|
|
web
|
snomed.info
|
131149001
|
|
web
|
snomed.info
|
131150001
|
|
web
|
snomed.info
|
131151002
|
|
web
|
snomed.info
|
131152009
|
|
web
|
snomed.info
|
131153004
|
|
web
|
snomed.info
|
131154005
|
|
web
|
snomed.info
|
131155006
|
|
web
|
snomed.info
|
131156007
|
|
web
|
snomed.info
|
131157003
|
|
web
|
snomed.info
|
131158008
|
|
web
|
snomed.info
|
131159000
|
|
web
|
snomed.info
|
131160005
|
|
web
|
snomed.info
|
131161009
|
|
web
|
snomed.info
|
131162002
|
|
web
|
snomed.info
|
131163007
|
|
web
|
snomed.info
|
131164001
|
|
web
|
snomed.info
|
131165000
|
|
web
|
snomed.info
|
131166004
|
|
web
|
snomed.info
|
131167008
|
|
web
|
snomed.info
|
131168003
|
|
web
|
snomed.info
|
131169006
|
|
web
|
snomed.info
|
131178000
|
|
web
|
snomed.info
|
131179008
|
|
web
|
snomed.info
|
131180006
|
|
web
|
snomed.info
|
131181005
|
|
web
|
snomed.info
|
165743006
|
|
web
|
snomed.info
|
165746003
|
|
web
|
snomed.info
|
165747007
|
|
web
|
snomed.info
|
199738000
|
|
web
|
snomed.info
|
250376006
|
|
web
|
snomed.info
|
250685002
|
|
web
|
snomed.info
|
250686001
|
|
web
|
snomed.info
|
250687005
|
|
web
|
snomed.info
|
250688000
|
|
web
|
snomed.info
|
250689008
|
|
web
|
snomed.info
|
250690004
|
|
web
|
snomed.info
|
250691000
|
|
web
|
snomed.info
|
250693002
|
|
web
|
snomed.info
|
250694008
|
|
web
|
snomed.info
|
250695009
|
|
web
|
snomed.info
|
250696005
|
|
web
|
snomed.info
|
250697001
|
|
web
|
snomed.info
|
250698006
|
|
web
|
snomed.info
|
264771009
|
|
web
|
snomed.info
|
278147001
|
|
web
|
snomed.info
|
278148006
|
|
web
|
snomed.info
|
278149003
|
|
web
|
snomed.info
|
278150003
|
|
web
|
snomed.info
|
278151004
|
|
web
|
snomed.info
|
278152006
|
|
web
|
snomed.info
|
278153001
|
|
web
|
snomed.info
|
278154007
|
|
web
|
snomed.info
|
302960008
|
|
web
|
snomed.info
|
312969002
|
|
web
|
snomed.info
|
325753009
|
|
web
|
snomed.info
|
326724004
|
|
web
|
snomed.info
|
327671006
|
|
web
|
snomed.info
|
365832006
|
|
web
|
snomed.info
|
365833001
|
|
web
|
snomed.info
|
405847005
|
|
web
|
snomed.info
|
405848000
|
|
web
|
snomed.info
|
405849008
|
|
web
|
snomed.info
|
405850008
|
|
web
|
snomed.info
|
405851007
|
|
web
|
snomed.info
|
405852000
|
|
web
|
snomed.info
|
405853005
|
|
web
|
snomed.info
|
405854004
|
|
web
|
snomed.info
|
405855003
|
|
web
|
snomed.info
|
405856002
|
|
web
|
snomed.info
|
405857006
|
|
web
|
snomed.info
|
405858001
|
|
web
|
snomed.info
|
405859009
|
|
web
|
snomed.info
|
405860004
|
|
web
|
snomed.info
|
405861000
|
|
web
|
snomed.info
|
405862007
|
|
web
|
snomed.info
|
405863002
|
|
web
|
snomed.info
|
405864008
|
|
web
|
snomed.info
|
405865009
|
|
web
|
snomed.info
|
405866005
|
|
web
|
snomed.info
|
405868006
|
|
web
|
snomed.info
|
405869003
|
|
web
|
snomed.info
|
405870002
|
|
web
|
snomed.info
|
405871003
|
|
web
|
snomed.info
|
405872005
|
|
web
|
snomed.info
|
405873000
|
|
web
|
snomed.info
|
405874006
|
|
web
|
snomed.info
|
405875007
|
|
web
|
snomed.info
|
405876008
|
|
web
|
snomed.info
|
405877004
|
|
web
|
snomed.info
|
405878009
|
|
web
|
snomed.info
|
405879001
|
|
web
|
snomed.info
|
405880003
|
|
web
|
snomed.info
|
405881004
|
|
web
|
snomed.info
|
405882006
|
|
web
|
snomed.info
|
405883001
|
|
web
|
snomed.info
|
405884007
|
|
web
|
snomed.info
|
405885008
|
|
web
|
snomed.info
|
405886009
|
|
web
|
snomed.info
|
405887000
|
|
web
|
snomed.info
|
405888005
|
|
web
|
snomed.info
|
405889002
|
|
web
|
snomed.info
|
405890006
|
|
web
|
snomed.info
|
405891005
|
|
web
|
snomed.info
|
405892003
|
|
web
|
snomed.info
|
405893008
|
|
web
|
snomed.info
|
405894002
|
|
web
|
snomed.info
|
405895001
|
|
web
|
snomed.info
|
405896000
|
|
web
|
snomed.info
|
405897009
|
|
web
|
snomed.info
|
405898004
|
|
web
|
snomed.info
|
405899007
|
|
web
|
snomed.info
|
405900002
|
|
web
|
snomed.info
|
405901003
|
|
web
|
snomed.info
|
412730000
|
|
web
|
snomed.info
|
412731001
|
|
web
|
snomed.info
|
412734009
|
|
web
|
snomed.info
|
412736006
|
|
web
|
snomed.info
|
412738007
|
|
web
|
snomed.info
|
412739004
|
|
web
|
snomed.info
|
413596002
|
|
web
|
snomed.info
|
430934002
|
|
web
|
snomed.info
|
445180002
|
|
web
|
snomed.info
|
445333001
|
|
web
|
snomed.info
|
471281007
|
|
web
|
snomed.info
|
471282000
|
|
web
|
snomed.info
|
702781009
|
|
web
|
snomed.info
|
702782002
|
|
web
|
snomed.info
|
702783007
|
|
web
|
snomed.info
|
705105000
|
|
web
|
snomed.info
|
709075008
|
|
web
|
snomed.info
|
710010005
|
|
web
|
snomed.info
|
710019006
|
|
web
|
snomed.info
|
719007008
|
|
web
|
snomed.info
|
733119003
|
|
web
|
snomed.info
|
733120009
|
|
web
|
snomed.info
|
738288005
|
|
web
|
snomed.info
|
738289002
|
|
web
|
snomed.info
|
738532000
|
|
web
|
snomed.info
|
738533005
|
|
web
|
snomed.info
|
738534004
|
|
web
|
snomed.info
|
738535003
|
|
web
|
snomed.info
|
738536002
|
|
web
|
snomed.info
|
738537006
|
|
web
|
snomed.info
|
738538001
|
|
web
|
snomed.info
|
738539009
|
|
web
|
snomed.info
|
738540006
|
|
web
|
snomed.info
|
738541005
|
|
web
|
snomed.info
|
738542003
|
|
web
|
snomed.info
|
738543008
|
|
web
|
snomed.info
|
738544002
|
|
web
|
snomed.info
|
738545001
|
|
web
|
snomed.info
|
738546000
|
|
web
|
snomed.info
|
738547009
|
|
web
|
snomed.info
|
738760000
|
|
web
|
snomed.info
|
738761001
|
|
web
|
snomed.info
|
738762008
|
|
web
|
snomed.info
|
738763003
|
|
web
|
snomed.info
|
738764009
|
|
web
|
snomed.info
|
738765005
|
|
web
|
snomed.info
|
738766006
|
|
web
|
snomed.info
|
738782007
|
|
web
|
snomed.info
|
738783002
|
|
web
|
snomed.info
|
738784008
|
|
web
|
snomed.info
|
738785009
|
|
web
|
snomed.info
|
738786005
|
|
web
|
snomed.info
|
738787001
|
|
web
|
snomed.info
|
738788006
|
|
web
|
snomed.info
|
738789003
|
|
web
|
snomed.info
|
738790007
|
|
web
|
snomed.info
|
739062009
|
|
web
|
snomed.info
|
739063004
|
|
web
|
snomed.info
|
739064005
|
|
web
|
snomed.info
|
739071000
|
|
web
|
snomed.info
|
739072007
|
|
web
|
snomed.info
|
772077005
|
|
web
|
snomed.info
|
772078000
|
|
web
|
snomed.info
|
772107008
|
|
web
|
snomed.info
|
772108003
|
|
web
|
snomed.info
|
772109006
|
|
web
|
snomed.info
|
772110001
|
|
web
|
snomed.info
|
781386002
|
|
web
|
snomed.info
|
787128002
|
|
web
|
snomed.info
|
787129005
|
|
web
|
snomed.info
|
787130000
|
|
web
|
snomed.info
|
787131001
|
|
web
|
snomed.info
|
787132008
|
|
web
|
snomed.info
|
787133003
|
|
web
|
snomed.info
|
787134009
|
|
web
|
snomed.info
|
787154005
|
|
web
|
snomed.info
|
787192006
|
|
web
|
snomed.info
|
787193001
|
|
web
|
snomed.info
|
787194007
|
|
web
|
snomed.info
|
787358002
|
|
web
|
snomed.info
|
787360000
|
|
web
|
snomed.info
|
787363003
|
|
web
|
snomed.info
|
787364009
|
|
web
|
snomed.info
|
787365005
|
|
web
|
snomed.info
|
787366006
|
|
web
|
snomed.info
|
787367002
|
|
web
|
snomed.info
|
787368007
|
|
web
|
snomed.info
|
787369004
|
|
web
|
snomed.info
|
787380004
|
|
web
|
snomed.info
|
787381000
|
|
web
|
snomed.info
|
787382007
|
|
web
|
snomed.info
|
787383002
|
|
web
|
snomed.info
|
787384008
|
|
web
|
snomed.info
|
787385009
|
|
web
|
snomed.info
|
787386005
|
|
web
|
snomed.info
|
787403004
|
|
web
|
snomed.info
|
787404005
|
|
web
|
snomed.info
|
787405006
|
|
web
|
snomed.info
|
787406007
|
|
web
|
snomed.info
|
787433006
|
|
web
|
snomed.info
|
787434000
|
|
web
|
snomed.info
|
787435004
|
|
web
|
snomed.info
|
792857006
|
|
web
|
snomed.info
|
890189007
|
|
web
|
snomed.info
|
1003612008
|
|
web
|
snomed.info
|
1003641004
|
|
web
|
snomed.info
|
1004155003
|
|
web
|
snomed.info
|
1010400009
|
|
web
|
snomed.info
|
1141749000
|
|
web
|
snomed.info
|
1142134000
|
|
web
|
snomed.info
|
1144910008
|
|
web
|
snomed.info
|
1144913005
|
|
web
|
snomed.info
|
1144930007
|
|
web
|
snomed.info
|
1156628004
|
|
web
|
snomed.info
|
1162602001
|
|
web
|
snomed.info
|
1179760003
|
|
web
|
snomed.info
|
1217473009
|
|
web
|
snomed.info
|
1217474003
|
|
web
|
snomed.info
|
1237618009
|
|
web
|
snomed.info
|
1254911003
|
|
web
|
snomed.info
|
71951000119107
|
|
web
|
snomed.info
|
85121000119109
|
|
web
|
snomed.info
|
204871000237101
|
|
web
|
snomed.info
|
204881000237104
|
|
web
|
snomed.info
|
204891000237102
|
|
web
|
snomed.info
|
204901000237101
|
|
web
|
snomed.info
|
248211000000106
|
|
web
|
snomed.info
|
248221000000100
|
|
web
|
snomed.info
|
302841000000105
|
|
web
|
snomed.info
|
912201000000104
|
|
web
|
snomed.info
|
1099611000119109
|
|
web
|
www.england.nhs.uk
|
See National genomic test directory for rare and inherited disease
on Genomic Test Directory
|
|
web
|
www.england.nhs.uk
|
See NHS England Genomic Test Directory
|
|
web
|
www.england.nhs.uk
|
Source: NHS England - North West Region
|
|
web
|
profiles.ihe.net
|
IHE Patient Encounter Management [ITI-31]
|
|
web
|
snomed.info
|
77386006
|
|
web
|
snomed.info
|
255407002
|
|
web
|
www.datadictionary.nhs.uk
|
NHS Data Model and Dictionary
ADMISSION METHOD
|
|
web
|
www.datadictionary.nhs.uk
|
NHS Data Model and Dictionary
ADMISSION SOURCE
|
|
web
|
www.datadictionary.nhs.uk
|
NHS Data Model and Dictionary
DESTINATION OF DISCHARGE
|
|
web
|
www.datadictionary.nhs.uk
|
NHS Data Model and Dictionary
DISCHARGE METHOD CODE
|
|
web
|
fhir.ch
|
Switzerland DocumentEntry.classCode
|
|
web
|
github.com
|
NHS England NRL - Record Category
|
|
web
|
snomed.info
|
721981007
|
|
web
|
snomed.info
|
721963009
|
|
web
|
snomed.info
|
734163000
|
|
web
|
snomed.info
|
371525003
|
|
web
|
snomed.info
|
422735006
|
|
web
|
snomed.info
|
371537001
|
|
web
|
snomed.info
|
419891008
|
|
web
|
fhir.ch
|
Switzerland DocumentEntry.mimeType
|
|
web
|
www.digihealthcare.scot
|
Digital Health and Care Scotland
(EH4001) CLINICAL DOCUMENT INDEXING STANDARDS
LA Labs
|
|
web
|
github.com
|
NHS England NRL -Record Type
|
|
web
|
snomed.info
|
4241000179101
|
|
web
|
snomed.info
|
721965002
|
|
web
|
snomed.info
|
163221000000102
|
|
web
|
snomed.info
|
927061000000101
|
|
web
|
snomed.info
|
149671000000105
|
|
web
|
snomed.info
|
820161000000108
|
|
web
|
snomed.info
|
820191000000102
|
|
web
|
snomed.info
|
824231000000100
|
|
web
|
snomed.info
|
820211000000103
|
|
web
|
snomed.info
|
823611000000107
|
|
web
|
snomed.info
|
820441000000103
|
|
web
|
snomed.info
|
820221000000109
|
|
web
|
snomed.info
|
829201000000105
|
|
web
|
snomed.info
|
820451000000100
|
|
web
|
snomed.info
|
820461000000102
|
|
web
|
snomed.info
|
820471000000109
|
|
web
|
snomed.info
|
823631000000104
|
|
web
|
snomed.info
|
820481000000106
|
|
web
|
snomed.info
|
820491000000108
|
|
web
|
snomed.info
|
823641000000108
|
|
web
|
snomed.info
|
24681000000104
|
|
web
|
snomed.info
|
824321000000109
|
|
web
|
snomed.info
|
823651000000106
|
|
web
|
snomed.info
|
823681000000100
|
|
web
|
snomed.info
|
823691000000103
|
|
web
|
snomed.info
|
823701000000103
|
|
web
|
snomed.info
|
824331000000106
|
|
web
|
snomed.info
|
824341000000102
|
|
web
|
snomed.info
|
25611000000107
|
|
web
|
snomed.info
|
823721000000107
|
|
web
|
snomed.info
|
823761000000104
|
|
web
|
snomed.info
|
909921000000109
|
|
web
|
snomed.info
|
823781000000108
|
|
web
|
snomed.info
|
1325651000000108
|
|
web
|
snomed.info
|
820241000000102
|
|
web
|
snomed.info
|
962381000000101
|
|
web
|
snomed.info
|
1219151000000104
|
|
web
|
snomed.info
|
25831000000103
|
|
web
|
snomed.info
|
416779005
|
|
web
|
snomed.info
|
820501000000102
|
|
web
|
snomed.info
|
820511000000100
|
|
web
|
snomed.info
|
820251000000104
|
|
web
|
snomed.info
|
823831000000103
|
|
web
|
snomed.info
|
823661000000109
|
|
web
|
snomed.info
|
823841000000107
|
|
web
|
snomed.info
|
725869001
|
|
web
|
snomed.info
|
1129271000000109
|
|
web
|
snomed.info
|
1363671000000109
|
|
web
|
snomed.info
|
1198111000000109
|
|
web
|
snomed.info
|
1076911000000107
|
|
web
|
snomed.info
|
4321000179101
|
|
web
|
snomed.info
|
1054291000000102
|
|
web
|
snomed.info
|
1054281000000104
|
|
web
|
snomed.info
|
4341000179107
|
|
web
|
snomed.info
|
1054181000000105
|
|
web
|
snomed.info
|
909871000000100
|
|
web
|
snomed.info
|
1054161000000101
|
|
web
|
snomed.info
|
824781000000106
|
|
web
|
snomed.info
|
824791000000108
|
|
web
|
snomed.info
|
820271000000108
|
|
web
|
snomed.info
|
1099461000000101
|
|
web
|
snomed.info
|
1325821000000102
|
|
web
|
snomed.info
|
185361000000102
|
|
web
|
snomed.info
|
826501000000100
|
|
web
|
snomed.info
|
824801000000107
|
|
web
|
snomed.info
|
24761000000103
|
|
web
|
snomed.info
|
823871000000101
|
|
web
|
snomed.info
|
229054004
|
|
web
|
snomed.info
|
824831000000101
|
|
web
|
snomed.info
|
307930005
|
|
web
|
snomed.info
|
826511000000103
|
|
web
|
snomed.info
|
820291000000107
|
|
web
|
snomed.info
|
826621000000105
|
|
web
|
snomed.info
|
826631000000107
|
|
web
|
snomed.info
|
826521000000109
|
|
web
|
snomed.info
|
308575004
|
|
web
|
snomed.info
|
310854009
|
|
web
|
snomed.info
|
308619006
|
|
web
|
snomed.info
|
270372007
|
|
web
|
snomed.info
|
270370004
|
|
web
|
snomed.info
|
307881004
|
|
web
|
snomed.info
|
270358003
|
|
web
|
snomed.info
|
308621001
|
|
web
|
snomed.info
|
820301000000106
|
|
web
|
snomed.info
|
823951000000100
|
|
web
|
snomed.info
|
826541000000102
|
|
web
|
snomed.info
|
826651000000100
|
|
web
|
snomed.info
|
408403008
|
|
web
|
snomed.info
|
823881000000104
|
|
web
|
snomed.info
|
827701000000106
|
|
web
|
snomed.info
|
772790007
|
|
web
|
snomed.info
|
822761000000108
|
|
web
|
snomed.info
|
1054091000000108
|
|
web
|
snomed.info
|
1054071000000109
|
|
web
|
snomed.info
|
827711000000108
|
|
web
|
snomed.info
|
822771000000101
|
|
web
|
snomed.info
|
822781000000104
|
|
web
|
snomed.info
|
822801000000103
|
|
web
|
snomed.info
|
822791000000102
|
|
web
|
snomed.info
|
827721000000102
|
|
web
|
snomed.info
|
822811000000101
|
|
web
|
snomed.info
|
824861000000106
|
|
web
|
snomed.info
|
371527006
|
|
web
|
snomed.info
|
822821000000107
|
|
web
|
snomed.info
|
822831000000109
|
|
web
|
snomed.info
|
823891000000102
|
|
web
|
snomed.info
|
962401000000101
|
|
web
|
snomed.info
|
4271000179106
|
|
web
|
snomed.info
|
983701000000101
|
|
web
|
snomed.info
|
725870000
|
|
web
|
snomed.info
|
725956001
|
|
web
|
snomed.info
|
770573007
|
|
web
|
snomed.info
|
770574001
|
|
web
|
snomed.info
|
770572002
|
|
web
|
snomed.info
|
770575000
|
|
web
|
snomed.info
|
782660000
|
|
web
|
snomed.info
|
782659005
|
|
web
|
snomed.info
|
1129261000000102
|
|
web
|
snomed.info
|
1129251000000100
|
|
web
|
snomed.info
|
1129241000000103
|
|
web
|
snomed.info
|
1129231000000107
|
|
web
|
snomed.info
|
4201000179104
|
|
web
|
snomed.info
|
1219161000000101
|
|
web
|
snomed.info
|
149691000000109
|
|
web
|
snomed.info
|
229059009
|
|
web
|
snomed.info
|
823901000000101
|
|
web
|
snomed.info
|
823931000000107
|
|
web
|
snomed.info
|
823941000000103
|
|
web
|
www.datadictionary.nhs.uk
|
NHS Data Model and Dictionary
ETHNIC CATEGORY
This extends UKCoreEthnicCategory
to include other categories for reasons stated in Wikipedia: Classification of ethnicity in the United Kingdom
|
|
web
|
en.wikipedia.org
|
NHS Data Model and Dictionary
ETHNIC CATEGORY
This extends UKCoreEthnicCategory
to include other categories for reasons stated in Wikipedia: Classification of ethnicity in the United Kingdom
|
|
web
|
www.datadictionary.nhs.uk
|
NHS Data Model and Dictionary
ACTIVITY LOCATION TYPE CODE
|
|
web
|
snomed.info
|
1186936003
|
|
web
|
www.datadictionary.nhs.uk
|
DRAFT
NHS Data Model and Dictionary
TREATMENT FUNCTION CODE
|
|
web
|
snomed.info
|
736622005
|
|
web
|
snomed.info
|
310000008
|
|
web
|
snomed.info
|
445449000
|
|
web
|
snomed.info
|
2421000175108
|
|
web
|
snomed.info
|
931781000000102
|
|
web
|
snomed.info
|
310072004
|
|
web
|
snomed.info
|
788125002
|
|
web
|
snomed.info
|
828821000000105
|
|
web
|
snomed.info
|
24271000087103
|
|
web
|
snomed.info
|
310013002
|
|
web
|
snomed.info
|
893391000000101
|
|
web
|
snomed.info
|
23871000087101
|
|
web
|
snomed.info
|
310006002
|
|
web
|
snomed.info
|
23891000087102
|
|
web
|
snomed.info
|
310016005
|
|
web
|
snomed.info
|
310033003
|
|
web
|
snomed.info
|
788002001
|
|
web
|
snomed.info
|
1362761000000103
|
|
web
|
snomed.info
|
840587001
|
|
web
|
snomed.info
|
711332004
|
|
web
|
snomed.info
|
310001007
|
|
web
|
snomed.info
|
708183009
|
|
web
|
snomed.info
|
827641000000101
|
|
web
|
snomed.info
|
310083008
|
|
web
|
snomed.info
|
310082003
|
|
web
|
snomed.info
|
91901000000109
|
|
web
|
snomed.info
|
310002000
|
|
web
|
snomed.info
|
310021008
|
|
web
|
snomed.info
|
310008001
|
|
web
|
snomed.info
|
310004004
|
|
web
|
snomed.info
|
1323641000000106
|
|
web
|
snomed.info
|
310011000
|
|
web
|
snomed.info
|
1323631000000102
|
|
web
|
snomed.info
|
706901001
|
|
web
|
snomed.info
|
2391000175104
|
|
web
|
snomed.info
|
408452007
|
|
web
|
snomed.info
|
892791000000108
|
|
web
|
snomed.info
|
708194008
|
|
web
|
snomed.info
|
932241000000105
|
|
web
|
snomed.info
|
310126000
|
|
web
|
snomed.info
|
310139001
|
|
web
|
snomed.info
|
24051000087102
|
|
web
|
snomed.info
|
892781000000106
|
|
web
|
snomed.info
|
395104009
|
|
web
|
snomed.info
|
1323651000000109
|
|
web
|
snomed.info
|
733459009
|
|
web
|
snomed.info
|
310142007
|
|
web
|
snomed.info
|
708172000
|
|
web
|
snomed.info
|
789718008
|
|
web
|
snomed.info
|
310140004
|
|
web
|
snomed.info
|
892771000000109
|
|
web
|
snomed.info
|
310117003
|
|
web
|
snomed.info
|
310003005
|
|
web
|
snomed.info
|
788127005
|
|
web
|
snomed.info
|
310099002
|
|
web
|
snomed.info
|
828811000000104
|
|
web
|
snomed.info
|
310104001
|
|
web
|
snomed.info
|
722170006
|
|
web
|
snomed.info
|
830038007
|
|
web
|
snomed.info
|
310076001
|
|
web
|
snomed.info
|
788004000
|
|
web
|
snomed.info
|
892761000000102
|
|
web
|
snomed.info
|
788121006
|
|
web
|
snomed.info
|
830037002
|
|
web
|
snomed.info
|
892741000000103
|
|
web
|
snomed.info
|
830149003
|
|
web
|
snomed.info
|
310022001
|
|
web
|
snomed.info
|
708184003
|
|
web
|
snomed.info
|
700436003
|
|
web
|
snomed.info
|
700435004
|
|
web
|
snomed.info
|
893381000000103
|
|
web
|
snomed.info
|
708193002
|
|
web
|
snomed.info
|
310012007
|
|
web
|
snomed.info
|
310155008
|
|
web
|
snomed.info
|
310024000
|
|
web
|
snomed.info
|
699650006
|
|
web
|
snomed.info
|
829981000000108
|
|
web
|
snomed.info
|
413294000
|
|
web
|
snomed.info
|
408451000
|
|
web
|
snomed.info
|
89301000000108
|
|
web
|
snomed.info
|
714089006
|
|
web
|
snomed.info
|
931801000000101
|
|
web
|
snomed.info
|
310108003
|
|
web
|
snomed.info
|
310111002
|
|
web
|
snomed.info
|
310067008
|
|
web
|
snomed.info
|
310131003
|
|
web
|
snomed.info
|
1240241000000109
|
|
web
|
snomed.info
|
395086005
|
|
web
|
snomed.info
|
310114005
|
|
web
|
snomed.info
|
310091004
|
|
web
|
snomed.info
|
310094007
|
|
web
|
snomed.info
|
310088004
|
|
web
|
snomed.info
|
310102002
|
|
web
|
snomed.info
|
310025004
|
|
web
|
snomed.info
|
893351000000109
|
|
web
|
snomed.info
|
310128004
|
|
web
|
snomed.info
|
893341000000106
|
|
web
|
snomed.info
|
310026003
|
|
web
|
snomed.info
|
89311000000105
|
|
web
|
snomed.info
|
788128000
|
|
web
|
snomed.info
|
700231006
|
|
web
|
snomed.info
|
708178001
|
|
web
|
snomed.info
|
310200001
|
|
web
|
snomed.info
|
310084002
|
|
web
|
snomed.info
|
893331000000102
|
|
web
|
snomed.info
|
828191000000104
|
|
web
|
snomed.info
|
892731000000107
|
|
web
|
snomed.info
|
828201000000102
|
|
web
|
snomed.info
|
310143002
|
|
web
|
snomed.info
|
722176000
|
|
web
|
snomed.info
|
700241009
|
|
web
|
snomed.info
|
708180007
|
|
web
|
snomed.info
|
444913002
|
|
web
|
snomed.info
|
734920002
|
|
web
|
snomed.info
|
963151000000104
|
|
web
|
snomed.info
|
310005003
|
|
web
|
snomed.info
|
708175003
|
|
web
|
snomed.info
|
310028002
|
|
web
|
snomed.info
|
431051000124102
|
|
web
|
snomed.info
|
310090003
|
|
web
|
snomed.info
|
828291000000109
|
|
web
|
snomed.info
|
310010004
|
|
web
|
snomed.info
|
705150003
|
|
web
|
snomed.info
|
310029005
|
|
web
|
snomed.info
|
310085001
|
|
web
|
snomed.info
|
310149003
|
|
web
|
snomed.info
|
92191000000105
|
|
web
|
snomed.info
|
413299005
|
|
web
|
snomed.info
|
828281000000107
|
|
web
|
snomed.info
|
700221004
|
|
web
|
snomed.info
|
828301000000108
|
|
web
|
snomed.info
|
792847005
|
|
web
|
snomed.info
|
409971007
|
|
web
|
snomed.info
|
2471000175109
|
|
web
|
snomed.info
|
310150003
|
|
web
|
snomed.info
|
700434000
|
|
web
|
snomed.info
|
734862008
|
|
web
|
snomed.info
|
310030000
|
|
web
|
snomed.info
|
310031001
|
|
web
|
snomed.info
|
1323431000000104
|
|
web
|
snomed.info
|
1326391000000100
|
|
web
|
snomed.info
|
310118008
|
|
web
|
snomed.info
|
983641000000106
|
|
web
|
snomed.info
|
700433006
|
|
web
|
snomed.info
|
1323561000000108
|
|
web
|
snomed.info
|
310151004
|
|
web
|
snomed.info
|
310144008
|
|
web
|
snomed.info
|
310152006
|
|
web
|
snomed.info
|
1323691000000101
|
|
web
|
snomed.info
|
700232004
|
|
web
|
snomed.info
|
788007007
|
|
web
|
snomed.info
|
310156009
|
|
web
|
snomed.info
|
788006003
|
|
web
|
snomed.info
|
830039004
|
|
web
|
snomed.info
|
24291000087104
|
|
web
|
snomed.info
|
3531000175102
|
|
web
|
snomed.info
|
892711000000104
|
|
web
|
snomed.info
|
310061009
|
|
web
|
snomed.info
|
708196005
|
|
web
|
snomed.info
|
893311000000105
|
|
web
|
snomed.info
|
310157000
|
|
web
|
snomed.info
|
310130002
|
|
web
|
snomed.info
|
1078501000000104
|
|
web
|
snomed.info
|
224891009
|
|
web
|
snomed.info
|
310015009
|
|
web
|
snomed.info
|
310020009
|
|
web
|
snomed.info
|
892621000000104
|
|
web
|
snomed.info
|
310158005
|
|
web
|
snomed.info
|
23901000087101
|
|
web
|
snomed.info
|
708182004
|
|
web
|
snomed.info
|
788124003
|
|
web
|
snomed.info
|
24081000087105
|
|
web
|
snomed.info
|
24101000087102
|
|
web
|
snomed.info
|
444933003
|
|
web
|
snomed.info
|
828331000000102
|
|
web
|
snomed.info
|
310107008
|
|
web
|
snomed.info
|
310110001
|
|
web
|
snomed.info
|
310113004
|
|
web
|
snomed.info
|
310092006
|
|
web
|
snomed.info
|
310096009
|
|
web
|
snomed.info
|
310098005
|
|
web
|
snomed.info
|
310089007
|
|
web
|
snomed.info
|
310103007
|
|
web
|
snomed.info
|
708190004
|
|
web
|
snomed.info
|
829951000000102
|
|
web
|
snomed.info
|
788001008
|
|
web
|
snomed.info
|
1323551000000105
|
|
web
|
snomed.info
|
1324191000000107
|
|
web
|
snomed.info
|
310032008
|
|
web
|
snomed.info
|
892601000000108
|
|
web
|
snomed.info
|
792848000
|
|
web
|
snomed.info
|
708174004
|
|
web
|
snomed.info
|
892581000000104
|
|
web
|
snomed.info
|
722393008
|
|
web
|
snomed.info
|
310119000
|
|
web
|
snomed.info
|
893301000000108
|
|
web
|
snomed.info
|
310127009
|
|
web
|
snomed.info
|
710028007
|
|
web
|
snomed.info
|
310078000
|
|
web
|
snomed.info
|
892571000000101
|
|
web
|
snomed.info
|
892561000000108
|
|
web
|
snomed.info
|
1323621000000104
|
|
web
|
snomed.info
|
373654008
|
|
web
|
snomed.info
|
893271000000105
|
|
web
|
snomed.info
|
23911000087104
|
|
web
|
snomed.info
|
310120006
|
|
web
|
snomed.info
|
310027007
|
|
web
|
snomed.info
|
92151000000102
|
|
web
|
snomed.info
|
893261000000103
|
|
web
|
snomed.info
|
92221000000103
|
|
web
|
snomed.info
|
893251000000101
|
|
web
|
snomed.info
|
708168004
|
|
web
|
snomed.info
|
714088003
|
|
web
|
snomed.info
|
708179009
|
|
web
|
snomed.info
|
310086000
|
|
web
|
snomed.info
|
706903003
|
|
web
|
snomed.info
|
706902008
|
|
web
|
snomed.info
|
23941000087103
|
|
web
|
snomed.info
|
24331000087108
|
|
web
|
snomed.info
|
310009009
|
|
web
|
snomed.info
|
893711000000109
|
|
web
|
snomed.info
|
741073001
|
|
web
|
snomed.info
|
840586005
|
|
web
|
snomed.info
|
788003006
|
|
web
|
snomed.info
|
788005004
|
|
web
|
snomed.info
|
310079008
|
|
web
|
snomed.info
|
1323921000000108
|
|
web
|
snomed.info
|
310159002
|
|
web
|
snomed.info
|
907301000000109
|
|
web
|
snomed.info
|
828511000000102
|
|
web
|
snomed.info
|
828521000000108
|
|
web
|
snomed.info
|
788009005
|
|
web
|
snomed.info
|
708170008
|
|
web
|
snomed.info
|
708173005
|
|
web
|
snomed.info
|
310060005
|
|
web
|
snomed.info
|
310063007
|
|
web
|
snomed.info
|
310064001
|
|
web
|
snomed.info
|
310093001
|
|
web
|
snomed.info
|
310065000
|
|
web
|
snomed.info
|
1323601000000108
|
|
web
|
snomed.info
|
310160007
|
|
web
|
snomed.info
|
23951000087100
|
|
web
|
snomed.info
|
310105000
|
|
web
|
snomed.info
|
788008002
|
|
web
|
snomed.info
|
931831000000107
|
|
web
|
snomed.info
|
931841000000103
|
|
web
|
snomed.info
|
931851000000100
|
|
web
|
snomed.info
|
310145009
|
|
web
|
snomed.info
|
310146005
|
|
web
|
snomed.info
|
1323571000000101
|
|
web
|
snomed.info
|
310161006
|
|
web
|
snomed.info
|
310106004
|
|
web
|
snomed.info
|
310109006
|
|
web
|
snomed.info
|
829961000000104
|
|
web
|
snomed.info
|
894001000000107
|
|
web
|
snomed.info
|
1323661000000107
|
|
web
|
snomed.info
|
893971000000101
|
|
web
|
snomed.info
|
893961000000108
|
|
web
|
snomed.info
|
789717003
|
|
web
|
snomed.info
|
24351000087104
|
|
web
|
snomed.info
|
792849008
|
|
web
|
snomed.info
|
893661000000101
|
|
web
|
snomed.info
|
893651000000104
|
|
web
|
snomed.info
|
1323821000000103
|
|
web
|
snomed.info
|
310014008
|
|
web
|
snomed.info
|
827981000000103
|
|
web
|
snomed.info
|
310147001
|
|
web
|
snomed.info
|
893941000000107
|
|
web
|
snomed.info
|
1079491000000102
|
|
web
|
snomed.info
|
893631000000106
|
|
web
|
snomed.info
|
310007006
|
|
web
|
snomed.info
|
3751000175100
|
|
web
|
snomed.info
|
3761000175103
|
|
web
|
snomed.info
|
893611000000103
|
|
web
|
snomed.info
|
3771000175106
|
|
web
|
snomed.info
|
893911000000106
|
|
web
|
snomed.info
|
897188002
|
|
web
|
snomed.info
|
310017001
|
|
web
|
snomed.info
|
1323851000000108
|
|
web
|
snomed.info
|
3781000175109
|
|
web
|
snomed.info
|
1323611000000105
|
|
web
|
snomed.info
|
310034009
|
|
web
|
snomed.info
|
893891000000108
|
|
web
|
snomed.info
|
893881000000106
|
|
web
|
snomed.info
|
893601000000100
|
|
web
|
snomed.info
|
893591000000106
|
|
web
|
snomed.info
|
3791000175107
|
|
web
|
snomed.info
|
893861000000102
|
|
web
|
snomed.info
|
310068003
|
|
web
|
snomed.info
|
893851000000100
|
|
web
|
snomed.info
|
310069006
|
|
web
|
snomed.info
|
893801000000101
|
|
web
|
snomed.info
|
1323801000000107
|
|
web
|
snomed.info
|
789716007
|
|
web
|
snomed.info
|
893791000000100
|
|
web
|
snomed.info
|
1323841000000105
|
|
web
|
snomed.info
|
24001000087103
|
|
web
|
snomed.info
|
3801000175108
|
|
web
|
snomed.info
|
893771000000104
|
|
web
|
snomed.info
|
789715006
|
|
web
|
snomed.info
|
789714005
|
|
web
|
snomed.info
|
310066004
|
|
web
|
snomed.info
|
310163009
|
|
web
|
snomed.info
|
893701000000107
|
|
web
|
snomed.info
|
893691000000107
|
|
web
|
snomed.info
|
893681000000105
|
|
web
|
snomed.info
|
893671000000108
|
|
web
|
snomed.info
|
310071006
|
|
web
|
snomed.info
|
310073009
|
|
web
|
snomed.info
|
893641000000102
|
|
web
|
snomed.info
|
310162004
|
|
web
|
snomed.info
|
706900000
|
|
web
|
snomed.info
|
310074003
|
|
web
|
snomed.info
|
901221000000102
|
|
web
|
snomed.info
|
1079481000000104
|
|
web
|
snomed.info
|
2451000175103
|
|
web
|
snomed.info
|
61831000000105
|
|
web
|
snomed.info
|
148581000000100
|
|
web
|
snomed.info
|
983341000000102
|
|
web
|
snomed.info
|
310080006
|
|
web
|
snomed.info
|
722424008
|
|
web
|
snomed.info
|
773558007
|
|
web
|
snomed.info
|
722140001
|
|
web
|
snomed.info
|
310164003
|
|
web
|
snomed.info
|
310100005
|
|
web
|
snomed.info
|
893231000000108
|
|
web
|
snomed.info
|
310087009
|
|
web
|
snomed.info
|
1325831000000100
|
|
web
|
snomed.info
|
840585009
|
|
web
|
snomed.info
|
310062002
|
|
web
|
snomed.info
|
278032008
|
|
web
|
snomed.info
|
310081005
|
|
web
|
snomed.info
|
828861000000102
|
|
web
|
snomed.info
|
816075004
|
|
web
|
snomed.info
|
788126001
|
|
web
|
snomed.info
|
734863003
|
|
web
|
snomed.info
|
893531000000105
|
|
web
|
snomed.info
|
310116007
|
|
web
|
snomed.info
|
310121005
|
|
web
|
snomed.info
|
310123008
|
|
web
|
snomed.info
|
722175001
|
|
web
|
snomed.info
|
310124002
|
|
web
|
snomed.info
|
932841000000106
|
|
web
|
snomed.info
|
310115006
|
|
web
|
snomed.info
|
722174002
|
|
web
|
snomed.info
|
2461000175101
|
|
web
|
snomed.info
|
788123009
|
|
web
|
snomed.info
|
310125001
|
|
web
|
snomed.info
|
310023006
|
|
web
|
snomed.info
|
1323901000000104
|
|
web
|
snomed.info
|
1323871000000104
|
|
web
|
snomed.info
|
310122003
|
|
web
|
snomed.info
|
310129007
|
|
web
|
snomed.info
|
908981000000101
|
|
web
|
snomed.info
|
816074000
|
|
web
|
snomed.info
|
893521000000108
|
|
web
|
snomed.info
|
708169007
|
|
web
|
snomed.info
|
893451000000102
|
|
web
|
snomed.info
|
310148006
|
|
web
|
snomed.info
|
3621000175101
|
|
web
|
snomed.info
|
1326421000000106
|
|
web
|
snomed.info
|
148621000000100
|
|
web
|
snomed.info
|
931821000000105
|
|
web
|
snomed.info
|
708188000
|
|
web
|
snomed.info
|
224930009
|
|
web
|
snomed.info
|
788122004
|
|
web
|
snomed.info
|
1323541000000107
|
|
web
|
snomed.info
|
310134006
|
|
web
|
snomed.info
|
310135007
|
|
web
|
snomed.info
|
310136008
|
|
web
|
snomed.info
|
310095008
|
|
web
|
snomed.info
|
310070007
|
|
web
|
snomed.info
|
1323501000000109
|
|
web
|
snomed.info
|
408458006
|
|
web
|
snomed.info
|
395092004
|
|
web
|
snomed.info
|
893221000000106
|
|
web
|
snomed.info
|
310101009
|
|
web
|
snomed.info
|
310018006
|
|
web
|
snomed.info
|
893211000000100
|
|
web
|
snomed.info
|
893121000000102
|
|
web
|
snomed.info
|
24141000087104
|
|
web
|
snomed.info
|
893201000000102
|
|
web
|
snomed.info
|
2351000175106
|
|
web
|
snomed.info
|
1323881000000102
|
|
web
|
snomed.info
|
310137004
|
|
web
|
snomed.info
|
109201000000109
|
|
web
|
snomed.info
|
310112009
|
|
web
|
snomed.info
|
699478002
|
|
web
|
snomed.info
|
708187005
|
|
web
|
snomed.info
|
310138009
|
|
web
|
snomed.info
|
310133000
|
|
web
|
snomed.info
|
911221000000100
|
|
web
|
snomed.info
|
911231000000103
|
|
web
|
snomed.info
|
911381000000108
|
|
web
|
snomed.info
|
310141000
|
|
web
|
snomed.info
|
310019003
|
|
web
|
snomed.info
|
708191000
|
|
web
|
snomed.info
|
896974005
|
|
web
|
snomed.info
|
893041000000108
|
|
web
|
snomed.info
|
733921009
|
|
web
|
snomed.info
|
310165002
|
|
web
|
snomed.info
|
893431000000109
|
|
web
|
snomed.info
|
310166001
|
|
web
|
snomed.info
|
893421000000107
|
|
web
|
snomed.info
|
310169008
|
|
web
|
snomed.info
|
310153001
|
|
web
|
snomed.info
|
1323531000000103
|
|
web
|
snomed.info
|
310167005
|
|
web
|
snomed.info
|
24011000087101
|
|
web
|
snomed.info
|
722352000
|
|
web
|
snomed.info
|
1323701000000101
|
|
web
|
snomed.info
|
310168000
|
|
web
|
snomed.info
|
708171007
|
|
web
|
snomed.info
|
708185002
|
|
web
|
snomed.info
|
413331009
|
|
web
|
snomed.info
|
893581000000109
|
|
web
|
snomed.info
|
828371000000100
|
|
web
|
snomed.info
|
828381000000103
|
|
web
|
snomed.info
|
310132005
|
|
web
|
www.datadictionary.nhs.uk
|
NHS Data Model and Dictionary
MAIN SPECIALTY CODE
|
|
web
|
snomed.info
|
408467006
|
|
web
|
snomed.info
|
394577000
|
|
web
|
snomed.info
|
394578005
|
|
web
|
snomed.info
|
421661004
|
|
web
|
snomed.info
|
408462000
|
|
web
|
snomed.info
|
394579002
|
|
web
|
snomed.info
|
394804000
|
|
web
|
snomed.info
|
394580004
|
|
web
|
snomed.info
|
394803006
|
|
web
|
snomed.info
|
408480009
|
|
web
|
snomed.info
|
408454008
|
|
web
|
snomed.info
|
394809005
|
|
web
|
snomed.info
|
394592004
|
|
web
|
snomed.info
|
394600006
|
|
web
|
snomed.info
|
394601005
|
|
web
|
snomed.info
|
394581000
|
|
web
|
snomed.info
|
408478003
|
|
web
|
snomed.info
|
394812008
|
|
web
|
snomed.info
|
408444009
|
|
web
|
snomed.info
|
394582007
|
|
web
|
snomed.info
|
408475000
|
|
web
|
snomed.info
|
410005002
|
|
web
|
snomed.info
|
394583002
|
|
web
|
snomed.info
|
419772000
|
|
web
|
snomed.info
|
394584008
|
|
web
|
snomed.info
|
408443003
|
|
web
|
snomed.info
|
394802001
|
|
web
|
snomed.info
|
394915009
|
|
web
|
snomed.info
|
394814009
|
|
web
|
snomed.info
|
394808002
|
|
web
|
snomed.info
|
394811001
|
|
web
|
snomed.info
|
408446006
|
|
web
|
snomed.info
|
394586005
|
|
web
|
snomed.info
|
394916005
|
|
web
|
snomed.info
|
408472002
|
|
web
|
snomed.info
|
394597005
|
|
web
|
snomed.info
|
394598000
|
|
web
|
snomed.info
|
394807007
|
|
web
|
snomed.info
|
419192003
|
|
web
|
snomed.info
|
408468001
|
|
web
|
snomed.info
|
394593009
|
|
web
|
snomed.info
|
394813003
|
|
web
|
snomed.info
|
410001006
|
|
web
|
snomed.info
|
394589003
|
|
web
|
snomed.info
|
394591006
|
|
web
|
snomed.info
|
394599008
|
|
web
|
snomed.info
|
394649004
|
|
web
|
snomed.info
|
408470005
|
|
web
|
snomed.info
|
394585009
|
|
web
|
snomed.info
|
394821009
|
|
web
|
snomed.info
|
422191005
|
|
web
|
snomed.info
|
394594003
|
|
web
|
snomed.info
|
416304004
|
|
web
|
snomed.info
|
418960008
|
|
web
|
snomed.info
|
394882004
|
|
web
|
snomed.info
|
394806003
|
|
web
|
snomed.info
|
394588006
|
|
web
|
snomed.info
|
408459003
|
|
web
|
snomed.info
|
394607009
|
|
web
|
snomed.info
|
419610006
|
|
web
|
snomed.info
|
418058008
|
|
web
|
snomed.info
|
420208008
|
|
web
|
snomed.info
|
418652005
|
|
web
|
snomed.info
|
418535003
|
|
web
|
snomed.info
|
418862001
|
|
web
|
snomed.info
|
419365004
|
|
web
|
snomed.info
|
418002000
|
|
web
|
snomed.info
|
419983000
|
|
web
|
snomed.info
|
419170002
|
|
web
|
snomed.info
|
419472004
|
|
web
|
snomed.info
|
394539006
|
|
web
|
snomed.info
|
420112009
|
|
web
|
snomed.info
|
409968004
|
|
web
|
snomed.info
|
394587001
|
|
web
|
snomed.info
|
394913002
|
|
web
|
snomed.info
|
408440000
|
|
web
|
snomed.info
|
418112009
|
|
web
|
snomed.info
|
419815003
|
|
web
|
snomed.info
|
394914008
|
|
web
|
snomed.info
|
408455009
|
|
web
|
snomed.info
|
394602003
|
|
web
|
snomed.info
|
408447002
|
|
web
|
snomed.info
|
394810000
|
|
web
|
snomed.info
|
408450004
|
|
web
|
snomed.info
|
408476004
|
|
web
|
snomed.info
|
408469009
|
|
web
|
snomed.info
|
408466002
|
|
web
|
snomed.info
|
408471009
|
|
web
|
snomed.info
|
408464004
|
|
web
|
snomed.info
|
408441001
|
|
web
|
snomed.info
|
408465003
|
|
web
|
snomed.info
|
394605001
|
|
web
|
snomed.info
|
394608004
|
|
web
|
snomed.info
|
408461007
|
|
web
|
snomed.info
|
408460008
|
|
web
|
snomed.info
|
394606000
|
|
web
|
snomed.info
|
408449004
|
|
web
|
snomed.info
|
418018006
|
|
web
|
snomed.info
|
394604002
|
|
web
|
snomed.info
|
394609007
|
|
web
|
snomed.info
|
408474001
|
|
web
|
snomed.info
|
394610002
|
|
web
|
snomed.info
|
394611003
|
|
web
|
snomed.info
|
408477008
|
|
web
|
snomed.info
|
394801008
|
|
web
|
snomed.info
|
408463005
|
|
web
|
snomed.info
|
419321007
|
|
web
|
snomed.info
|
394576009
|
|
web
|
snomed.info
|
394590007
|
|
web
|
snomed.info
|
409967009
|
|
web
|
snomed.info
|
408448007
|
|
web
|
snomed.info
|
419043006
|
|
web
|
snomed.info
|
394612005
|
|
web
|
snomed.info
|
394733009
|
|
web
|
snomed.info
|
394732004
|
|
web
|
snomed.info
|
119373006
|
|
web
|
snomed.info
|
309201001
|
|
web
|
snomed.info
|
258580003
|
|
web
|
snomed.info
|
122552005
|
|
web
|
snomed.info
|
122556008
|
|
web
|
snomed.info
|
737357006
|
|
web
|
snomed.info
|
122555007
|
|
web
|
snomed.info
|
440500007
|
|
web
|
snomed.info
|
119359002
|
|
web
|
snomed.info
|
733104004
|
|
web
|
snomed.info
|
258450006
|
|
web
|
snomed.info
|
258565009
|
|
web
|
snomed.info
|
702451000
|
|
web
|
snomed.info
|
309147000
|
|
web
|
snomed.info
|
258566005
|
|
web
|
snomed.info
|
441652008
|
|
web
|
snomed.info
|
1003517007
|
|
web
|
snomed.info
|
122571007
|
|
web
|
snomed.info
|
418564007
|
|
web
|
snomed.info
|
119342007
|
|
web
|
snomed.info
|
441479001
|
|
web
|
snomed.info
|
122575003
|
|
web
|
snomed.info
|
441673008
|
|
web
|
en.wikipedia.org
|
The architecture generally follows Domain Driven Design [DDD]
, Domain Driven Design
and Data Mesh
|
|
web
|
martinfowler.com
|
The architecture generally follows Domain Driven Design [DDD]
, Domain Driven Design
and Data Mesh
|
|
web
|
en.wikipedia.org
|
The architecture generally follows Domain Driven Design [DDD]
, Domain Driven Design
and Data Mesh
|
|
web
|
en.wikipedia.org
|
The Intermediary
, North West GMSA Regional Integration Engine (RIE) is an Enterprise Service Bus
most commonly known in the NHS as a Trust Integration Engine (TIE).
|
|
web
|
www.enterpriseintegrationpatterns.com
|
This implement as series of Enterprise Integration Patterns
based around messaging, the diagrams below follow conventions used for these patterns.
|
|
web
|
www.enterpriseintegrationpatterns.com
|
The ESB has a Canonical Data Model
which is expressed in this Implementation Guide using HL7 FHIR. This model is common to all the exchange formats used in the ESB:
|
|
web
|
en.wikipedia.org
|
pipe+hat HL7 v2
|
|
web
|
profiles.ihe.net
|
potential use case for sharing reports: XML IHE XDS.b
|
|
web
|
nw-gmsa.github.io
|
Digital Health and Care Wales - HL7 ORU_R01 2.5.1 Implementation Guide
|
|
web
|
www.rcr.ac.uk
|
Royal College of Radiologist
|
|
web
|
www.ihe-europe.net
|
IHE Europe Metadata for exchange medical documents and images
see UK content.
|
|
web
|
www.datadictionary.nhs.uk
|
NHS Data Model and Dictionary
|
|
web
|
www.enterpriseintegrationpatterns.com
|
This canonical model is not specific to Genomics. It is focused on standard message construction patterns in particular CorrelationIdentifier
such as Order Numbers and Episode/Stay Identifiers and use of Clinical Coding Systems such as UK SNOMED CT.
|
|
web
|
wiki.ihe.net
|
To support genomics workflow, this guide is aligned to enterprise workflow processes described in IHE Laboratory Testing Workflow
, terminology from this guide especially around Actors is used throughout this Implementation Guide.
|
|
web
|
www.enterpriseintegrationpatterns.com
|
First point of entry for HL7 FHIR O21 messages Command Message
.
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Call NHS England PDS & Enrich Content ( Content Enricher
)
- Stores and enhances the message with additional data elements (GP Practice and ICS).
- Ensures only traced NHS Numbers are present in the message.
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Update Genomic Data Repository ( Wire Tap
)
-
Updates internal genomic data repository using FHIR RESTful interactions. ( Messaging Gateway
)
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Updates internal genomic data repository using FHIR RESTful interactions. ( Messaging Gateway
)
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Router ( Message Router
)
- Routes messages based on order metadata.
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Transform to HL7 v2 Message ( Message Translator
and v2 Canoncial Model
)
- Converts HL7 FHIR O21 messages into HL7 v2.5.1 OML_O21 format.
- These transformed messages are sent to NW GMSA LIMS iGene.
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Transform to HL7 v2 Message ( Message Translator
and v2 Canoncial Model
)
- Converts HL7 FHIR O21 messages into HL7 v2.5.1 OML_O21 format.
- These transformed messages are sent to NW GMSA LIMS iGene.
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Genomic Order Management Adaptor Service FHIR API ( Messaging Gateway
)
- Targets NHS England Genomic Order Management Service FHIR API which is the interface to external GMSA.
- This uses a FHIR RESTful API, similar to the Clinical Data Repository Adaptor, and like this service, the business logic (how to update the repository) is held within Regional Integrations Engine and this is not exposed externally.
|
|
web
|
www.enterpriseintegrationpatterns.com
|
NW GMSA LIMS (iGene) ( Document Message
)
- Produces genomic test results in HL7 v2.3 ORU_R01 messages.
- These are sent into the Enterprise Service Bus (ESB).
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Transform to HL7 FHIR Message ( Message Translator
and FHIR Canoncial Model
)
- Converts HL7 v2.3 message into a modern HL7 FHIR R01 message.
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Transform to HL7 FHIR Message ( Message Translator
and FHIR Canoncial Model
)
- Converts HL7 v2.3 message into a modern HL7 FHIR R01 message.
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Update Genomic Data Repository & Enrich Content ( Content Enricher
)
- Stores and enhances the message with additional data elements.
- Provides a consistent, enriched dataset for downstream use.
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Reports are sent to the NHS Trust which ordered the test ( Message Router
)
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Reports are sent to NHS ICS Health Information Exchange (HIE) for sharing the reports within the ICS, this is based on the GP Surgery for the patient which is obtained via a PDS lookup. ( Dynamic Router
)
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Transform to HL7 v2 Message ( Message Translator
and v2 Canoncial Model
)
- Converts enriched content back into a structured HL7 v2.x format for downstream systems that still rely on v2.
- This ensures backward compatibility with existing hospital systems.
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Transform to HL7 v2 Message ( Message Translator
and v2 Canoncial Model
)
- Converts enriched content back into a structured HL7 v2.x format for downstream systems that still rely on v2.
- This ensures backward compatibility with existing hospital systems.
|
|
web
|
www.enterpriseintegrationpatterns.com
|
A dedicated Repository Service captures and stores all enriched FHIR data. ( Messaging Gateway
)
- FHIR Repository Adapter converts incoming HL7 FHIR messages into a format suitable for storage.
- Data is stored in the Clinic Data Repository (IRIS FHIR Repository).
- Access is available via HL7 FHIR RESTful API.
|
|
web
|
digital.nhs.uk
|
NHS England - Application-restricted APIs
|
|
web
|
termbrowser.nhs.uk
|
1022491000000106 and 1491000237105 SNOMED UK Edition
|
|
web
|
profiles.ihe.net
|
Is based on IHE Internet User Authorization (IUA)
but using client-credentials
grant only (at present).
|
|
web
|
en.wikipedia.org
|
In software design, these areas are often referred to as domains
. The Genomic Diagnostic Workflow
operates across several of these domains — in software architecture terms, this is known as a bounded context
.
|
|
web
|
martinfowler.com
|
In software design, these areas are often referred to as domains
. The Genomic Diagnostic Workflow
operates across several of these domains — in software architecture terms, this is known as a bounded context
.
|
|
web
|
en.wikipedia.org
|
Health Informatics Archetype
, this is model associated with messaging.
|
|
web
|
en.wikipedia.org
|
Entities
such as HL7 v2 segments and FHIR resources.
|
|
web
|
en.wikipedia.org
|
Events
such as HL7 v2 ADT and SET event feeds.
|
|
web
|
en.wikipedia.org
|
The Archetype
follows the Domain Archetype
concept from Data Mesh
principles and serves as a bridge between data architecture and software engineering.
|
|
web
|
openehr.org
|
Although this guide primarily uses HL7 FHIR and HL7 v2 to support interactions between health providers, it does not mandate or promote the use of these technical standards for data persistence within EHR, LIMS, or analytics systems. Suppliers may adopt alternative standards where more appropriate, such as openEHR
for electronic health record systems or the Global Alliance for Genomics and Health (GA4GH)
standards for research and system databases.
|
|
web
|
www.ga4gh.org
|
Although this guide primarily uses HL7 FHIR and HL7 v2 to support interactions between health providers, it does not mandate or promote the use of these technical standards for data persistence within EHR, LIMS, or analytics systems. Suppliers may adopt alternative standards where more appropriate, such as openEHR
for electronic health record systems or the Global Alliance for Genomics and Health (GA4GH)
standards for research and system databases.
|
|
web
|
ckm.openehr.org
|
EPR (Electronic Patient Record) systems (e.g. openEHR Genomics
)
|
|
web
|
www.ga4gh.org
|
Genomic Applications (e.g. Global Alliance for Genomics and Health
)
|
|
web
|
www.rcr.ac.uk
|
Laboratory reporting:
Royal College of Radiologists HL7 Guide
|
|
web
|
drive.google.com
|
Patient administration:
NHS England HL7 v2 ADT Message Specification
|
|
web
|
www.ihe-europe.net
|
Document metadata:
IHE Europe Document Metadata Guidelines
|
|
web
|
www.digihealthcare.scot
|
Document type classification:
Digital Health and Care Scotland (EH4001) Clinical Document Indexing Standards
|
|
web
|
wiki.ihe.net
|
See IHE Specimen Event Tracking
(HL7 v2 SET)
|
|
web
|
profiles.ihe.net
|
See IHE MHD ITI-105
|
|
web
|
github.com
|
Requirements or issues are logged in the NH Genomics IG issues
|
|
web
|
open.epic.com
|
EPIC HL7 v2
See Outgoing Ancillary Orders
(EPR to RIE)
|
|
web
|
ehr.meditech.com
|
MEDITECH HL7 v2
|
|
web
|
open.epic.com
|
EPIC HL7 v2
See Discrete Genomic Results
(RIE to EPIC EPR)
|
|
web
|
profiles.ihe.net
|
IHE XDS Cross-Enterprise Document Sharing (XDS.b) or Cross-Enterprise Document Reliable Interchange (XDR) - Provide and Register Document Set-b [ITI-41]
|
|
web
|
profiles.ihe.net
|
IHE MHD Mobile access to Health Documents (MHD) - Simplified Publish [ITI-105]
|
|
web
|
open.epic.com
|
EPIC Incoming Scanned Document Link Interface Technical Specification
|
|
web
|
drive.google.com
|
This is based on the definition of MSH from NHS England HL7 v2 ADT Message Specification
.
|
|
web
|
drive.google.com
|
This is based on the definition of PID from NHS England HL7 v2 ADT Message Specification
and Digital Health and Care Wales - HL7 ORU_R01 2.5.1 Implementation Guide
|
|
web
|
drive.google.com
|
This is based on the definition of PV1 from NHS England HL7 v2 ADT Message Specification
|
|
web
|
drive.google.com
|
This is based on the definition of PL from NHS England HL7 v2 ADT Message Specification
SHOULD
be followed and SHALL
be used in ORC-12.
In addition, this includes of PL.11 to hold organisation ODS code.
|
|
web
|
www.rcr.ac.uk
|
This is based on the definitions of NDL from Royal College of Radiologists
|
|
web
|
drive.google.com
|
Extended Composite ID Number and Name for Persons.
The definition of XCN from NHS England HL7 v2 ADT Message Specification
SHOULD
be followed and SHALL
be used in ORC-12.
|
|
web
|
drive.google.com
|
Extended Composite Name and Identification Number for Organizations.
The definition of XON from NHS England HL7 v2 ADT Message Specification
SHOULD
be followed and SHALL
be used in ORC-21.
|
|
web
|
github.com
|
Original Hl7 v2 OML_O21
|
|
web
|
github.com
|
FHIR Message O21
|
|
web
|
github.com
|
North West Genomics HL7 v2 OML_O21
|
|
web
|
github.com
|
Original Hl7 v2 ORU_R01
|
|
web
|
github.com
|
FHIR Message R01
|
|
web
|
github.com
|
North West Genomics HL7 v2 ORU_R01
|
|
web
|
github.com
|
FHIR Message T02
|
|
web
|
github.com
|
North West Genomics HL7 v2 MDM_T02
|
|
web
|
www.digihealthcare.scot
|
Digital Health and Care Scotland - (EH4001) CLINICAL DOCUMENT INDEXING STANDARDS
|
|
web
|
www.ihe-europe.net
|
IHE Europe Document Metadata
(this contains references to NHS England Data Dictionary and Model)
|
|
web
|
www.enterpriseintegrationpatterns.com
|
Both guides are effectively merged in this guide and applied to both HL7 v2 and FHIR. The pattern used here is Canonical Data Mode
, this can also be applied to other formats such as IHE XDS and DICOM.
|
|
web
|
fhir.epic.com
|
For NHS trusts with EPRs with FHIR REST APIs (e.g. EPIC
, Oracle and Cerner), the FHIR system used here SHOULD
be the same.
An OID can be used instead of a FHIR system, this may be defined for use with IHE XDS.
|
|
web
|
www.enterpriseintegrationpatterns.com
|
The diagram below illustrates point to point
messaging between an Order Placer
and an Order Filler
. The Order Filler
is typically a Laboratory Information Management System (LIMS), while the Order Placer
is usually a clinical system such as an Electronic Patient Record (EPR).
|
|
web
|
profiles.ihe.net
|
IHE Basic Audit Log Patterns (BALP)
HL7 FHIR
|
|
web
|
www.enterpriseintegrationpatterns.com
|
This represents a shift from a message-oriented workflow (see EIP Messaging Patterns
) to a conversation-based workflow (see EIP Conversation Patterns
). Rather than relying on single, transactional messages, the workflow is managed as an ongoing conversation between participants.
|
|
web
|
www.enterpriseintegrationpatterns.com
|
This represents a shift from a message-oriented workflow (see EIP Messaging Patterns
) to a conversation-based workflow (see EIP Conversation Patterns
). Rather than relying on single, transactional messages, the workflow is managed as an ongoing conversation between participants.
|
|
web
|
digital.nhs.uk
|
It is anticipated that event notifications will be implemented using FHIR Subscriptions
, which support a publish–subscribe (pub/sub) pattern, or alternatively through the NHS England Multicast Notification Service API
.
|
|
web
|
wiki.ihe.net
|
Genomic diagnostic testing follows the same standardized process defined by the IHE Laboratory Testing Workflow
used in traditional laboratory testing.
This workflow has been enhanced to support the sharing of laboratory reports (documents) through Integrated Care Systems (ICS). In addition, a new mechanism for sharing laboratory reports has been introduced to establish a regional genomic data repository.
|
|
web
|
profiles.ihe.net
|
IHE Mobile Care Services Discovery (mCSD)
|
|
web
|
digital.nhs.uk
|
Organisation Data Service (ODS) Organisation Data Terminology - FHIR API
|
|
web
|
profiles.ihe.net
|
Pattern: FHIR RESTful + IHE Mobile access to Health Documents (MHD)
|
|
web
|
profiles.ihe.net
|
Pattern: FHIR RESTful + IHE Query for Existing Data for Mobile (QEDm)
also following HL7 Genomic Report
|
|
web
|
profiles.ihe.net
|
Pattern: FHIR RESTful + IHE Mobile Health Document Sharing
- this is essentially the same as the local genomic reports but the registry is now a seperate service.
|
|
web
|
profiles.ihe.net
|
Pattern: FHIR RESTful + IHE Mobile Health Document Sharing
- as above but the document is now a FHIR Document, probably HL7 Europe Laboratory Report
combined with HL7 Genomic Report
|
|
web
|
profiles.ihe.net
|
Pattern: FHIR RESTful + IHE Document Subscription for Mobile (DSUBm)
|
|
web
|
profiles.ihe.net
|
For local this would be using an OAuth2 Authorisation flow - see Authorisation (OAuth2)
In addition all queries would be audited e.g. follow IHE Basic Audit Log Patterns (BALP)
|
|
web
|
developer.nhs.uk
|
For national, access to local repositories would be required would again use IHE BALP. The authentication is likely to be SSP Retrieval
, this means the consumer does not talk directly to the repository.
|
|
web
|
github.com
|
This implementation guide is built using HL7 igPublisher
which has a guide Guidance for FHIR IG Creation
It is hosted on GitHub LTW Genomics
|
|
web
|
github.com
|
issues can be raised via GitHub Issues
|
|
web
|
github.com
|
changes can be raised via GitHub Pull requests
|
|
web
|
developer.community.nhs.uk
|
Issues and questions can be raised on NHS England Developer Community
|
|
web
|
digital.nhs.uk
|
All test patients (with a NHS Number) are on NHS England Personal Demographics Service - FHIR API
(Int environment).
|
|
web
|
www.odsdatasearchandexport.nhs.uk
|
P84673
|
|
web
|
www.odsdatasearchandexport.nhs.uk
|
N82035
|
|
web
|
www.odsdatasearchandexport.nhs.uk
|
P81002
|
|
web
|
www.odsdatasearchandexport.nhs.uk
|
B86016
|
|
web
|
www.odsdatasearchandexport.nhs.uk
|
F83004
|
|
web
|
www.odsdatasearchandexport.nhs.uk
|
M85124
|
|
web
|
www.odsdatasearchandexport.nhs.uk
|
Future: W91022
Current: N81082
|
|
web
|
www.odsdatasearchandexport.nhs.uk
|
Future: W91022
Current: N81082
|
|
web
|
www.odsdatasearchandexport.nhs.uk
|
C81010
|
|
web
|
www.odsdatasearchandexport.nhs.uk
|
N81027
|
|
web
|
www.odsdatasearchandexport.nhs.uk
|
N81087
|
|
web
|
www.odsdatasearchandexport.nhs.uk
|
C81065
|
|
web
|
www.odsdatasearchandexport.nhs.uk
|
B82045
|
|
web
|
www.odsdatasearchandexport.nhs.uk
|
A82025
|
|
web
|
www.odsdatasearchandexport.nhs.uk
|
A82038
|
|
web
|
www.odsdatasearchandexport.nhs.uk
|
Y02657
|
|
web
|
www.odsdatasearchandexport.nhs.uk
|
Y00206
|
|
web
|
digital.nhs.uk
|
The APIs are available on the Health and Social Care Network (HSCN)
|
|
web
|
10.165.194.216
|
See Placer Order Management [LAB-1]
and CapabilityStatement
|
|
web
|
10.165.194.217
|
See Placer Order Management [LAB-1]
and CapabilityStatement
|
|
web
|
www.oauth.com
|
Integration Testing - OAuth2 client-credentials
|